Avicenna J Dent Res. 13(4):142-147.
doi: 10.34172/ajdr.2021.27
Original Article
In Vitro and In Silico Evaluation of Biological Properties of Some 1, 3, 4-Oxadiazole Derivatives Against Streptococcus mutans and Their Interaction With Gbp-C by Molecular Docking
Behin Omidi 1  , Yasin SarveAhrabi 1, *
, Yasin SarveAhrabi 1, * 
Author information:
1Department of Biology, Central Tehran Branch, Islamic Azad University, Tehran, Iran.
*
Correspondence to Yasin SarveAhrabi, Ph.D. in Microbiology, Department of Biology, Central Tehran Branch, Islamic Azad University, Tehran, Iran, Tel: +989141483263, Email:
yasin.ahrabi2016@gmail.com
Abstract
Background: The need to replace new drug structures for the treatment of resistant strains has become essential. Streptococcus mutans is one of the most important factors in causing tooth decay. Glucan binding protein-C (Gbp-C) is a crucial mobileular floor protein that is worried in biofilm formation, and 1, 3, 4-oxadiazoles are new antibacterial structures. Accordingly, this study focused on assessing in vitro and in silico activity of our previously synthesized compounds of 1, 3, 4-oxadiazole against S. mutans.
Methods: To this end, our previously synthesized derivatives were re-synthesized and prepared, and then antibacterial susceptibility tests were used for inhibition zone, minimum inhibitory concentration (MIC), and minimum bactericidal concentration (MBC) test values. The molecular docking method was also applied to confirm the effect of compounds in interaction with the Gbp-C of S. mutans.
Results: All compounds showed different effects against the bacterial sample. Among these, the most effective ones were related to naphthalene (4d), fluorophenyl (4e), and dimethoxyphenyl (4h) derivatives against S. mutans, respectively. Other compounds also had antibacterial properties but to a lesser extent. In the molecular part, compounds 4d and 4h had the highest affinity to inhibit the GbpC-protein. compound 4d with amino acids ASP and GLN established 402 and 391 hydrogen bonds, respectively, and compound 4h with amino acids SER, GLU, THR, and TRP established 347, 360, 449, and 451 hydrogen bonds, respectively.
Conclusions: In general, 1, 3, 4-oxadiazoles containing naphthalene and dimethoxy phenyl functional groups in high concentrations can be good alternatives to the existing drugs for eliminating caries-causing tooth mutants that have drug resistance. It seems that more inhibitory effects can be observed on clinical specimens by adding different purposeful groups and increasing the destructive power of oxadiazole-based compounds.
Keywords: Oxadiazoles, Streptococcus mutans, Antibacterial activity, Molecular docking
Copyright and License Information
© 2021 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Citation: Omidi B, Yasin SarveAhrabi Y. In vitro and in silico evaluation of biological properties of some 1, 3, 4-oxadiazole derivatives against streptococcus mutans and their interaction with Gbp-c by molecular docking. Avicenna J Dent Res. 2021;13(4):142-147. doi: 10.34172/ajdr.2021.27.
Background
Highlights
Tooth decay is one of the maximum essential public fitness issues in numerous societies, especially in underdeveloped countries, and one of the maximum not unusual place sicknesses in evolved countries (1). This disease is a multifactorial and important complication that accounts for a large part of public health research, particularly microbiological research (2). Despite the influence of various factors in the development and progression of tooth decay, the activity of acid-producing bacteria, especially Streptococcus mutans biofilms, is the main cause of this complication (3). S. mutans is a gram-positive coccus and optional anaerobic. In addition, it is the most important and first microorganism whose carcinogenicity has been proven to be present in plaque. Overall, 25 species of S. mutans have so far been discovered that destruct tooth enamel by producing lactic acid and fermenting sucrose (4). This bacterium produces acid by metabolizing sugars in its environment, which eventually dissolves tooth enamel and causes decay (5). Biofilm is a complicated composed of microorganisms that can be connected to every different surface. Biofilm formation calls for the preliminary attachment of microorganisms to a surface, accompanied by bacterial overgrowth (6). The microorganisms making up the biofilm are mainly S. mutans and anaerobic bacteria with a composition that varies in different parts of the mouth. The formation of the biofilms of bacterium depends on the presence of a special enzyme called glucansucrase or glucosyltransferase (7). This enzyme is a member of the glycoside hydrolase enzyme family called GH70 and has different numbers of amino acids in various strains. Nonetheless, on average, it has 1700-1100 amino acids and is encoded by the GTF gene (8). Glucan sucrase synthesizes a special homopolymer called glucan by binding glucose units through various alpha bonds. The secretion of this viscous hemopolysaccharide by S. mutans causes the formation of the biofilms of this bacterium and their strong attachment to the tooth surface. S. mutans typically contains three types of glucan sucrase enzymes called GTF-B, GTF-C, and GTF-D. These enzymes are encoded by the genes of the same name, gene gtf-B and gtf-D by binding sugar units through alpha (1→3) and alpha(1→6) bonds, synthesize soluble and insoluble types of glucan hemopolysaccharide, but gene gtf-C, synthesizes both soluble and insoluble Glucan sucrase(9). It should be noted that 1, 3, 4-oxadiazoles are biologically energetic natural compounds. 1, 3, 4-oxadiazole is nitrogen- and oxygen-containing heterocycle, and one of the four isomers of oxadiazoles (10). Although 1, 3, 4-oxadiazole is not normally utilized in chemistry, its different compounds are significant (11). For instance, raltegravir is an HIV drug that includes a 1, 3, 4-oxadiazole ring (12). Other pharmaceutical tablets containing the 1, 3, 4-oxadiazole ring consist of fenadiazole, zibotentan, and doxazosin (13). According to these drugs and a wide range of biological activities of oxadiazole compounds, synthesizing new derivatives of 1, 3, 4-oxadiazole with antibacterial, antifungal, anti-cancer, antioxidant, antimalarial, and anti-inflammatory properties are important (14). In the present study, our previous 1, 3, 4-oxadiazole derivatives were re-synthesized, and the inhibitory effect of these derivatives against S. mutans and glucan binding protein-C (Gbp-C), as the most important regulatory protein in the bacterial cohesion and pathogenesis of this bacterium was investigated experimentally and computationally.
Materials and Methods
All starting Materials had been organized from Merck (Germany) and used shifting ahead with no greater filtration. The bacterial strain (S. mutans ATCC 35668) prepared from the Iranian Industrial Microorganisms Collection Center (Lyophilized). Microbiological assessments were accomplished using a Memmert-INC153T2T3 incubator. The crystallographic structure of the Gbp-C of S. mutans by the 6CAM code was downloaded from https://www.rcsb.org.
Chemical
Preparation of Derivatives
Our previously synthesized derivatives were re-synthesized according to (15).
Preparation of Compound Concentrations
Dimethyl sulfoxide (99%, DMSO-Merck, Germany) was used to dissolve all compounds. Initially, a concentration of 0.5 mg/mL was prepared from the powders of synthesized compounds and control samples (1:9 ratios). Afterward, they were kept at -18 ̊C in sterile test tubes until performing the tests.
Antibacterial Activity
Preparation of Bacterial Suspension
First, the lipophilic ampoule containing the S. mutans strain was opened under sterile conditions and transferred to the nutrient broth culture medium and incubated for 24 hours at 37̊C. Then, a linear culture was performed on the selective-differential culture medium and incubated for 24 hours at 37̊C to ensure that the bacteria were sheer in the nutrient broth medium (NB, Merck, Germany). Using a sampler, 1 mL of a 24-hour culture of microbial suspension was transferred to a tube containing sterile NB, and then the turbidity of the microbial suspension was visually compared to the McFarland standard set with a spectrophotometer at 625 nm and an absorption rate of 1.5 × 108 CFU/mL. The Mueller-Hinton agar (MHA) (Merck, Germany) culture medium was applied for the agar well diffusion test, and the Mueller-Hinton broth (MHB, Merck, Germany) culture medium was used to test the dilution in the tubes. All cultivation environments were organized in line with the manufacturer’s commands and sterilized the use of autoclaves.
Agar Well Diffusion Method
To perform this experiment, wells of 5 mm in diameter were created by a sterile pipette in MHA culture media containing cultured bacterial suspension. The wells were then filled with synthesized compounds (4a-4h) and positive control (Vancomycin) samples and put inside the incubator for 24 hours at 37̊C. It is worth noting that all the steps were performed near the flame and in a sterile environment (15).This experiment was repeated three times, and their mean was reported in the results.
Broth Dilution Method
To determine the minimum inhibitory concentration (MIC), a series of 9 tubes were employed to test the different dilutions of each compound. It is noteworthy that the control sample (vancomycin) was diluted in 9 separate tubes. The initial concentration of each compound was 2 mg/mL, which was obtained by inserting 1 mL of the compound into the first tube containing 1 mL of culture medium at a concentration of 1 mg/mL. Different dilutions were obtained from tube number 1 (2 mg/mL) to tube number 9 (0.007 mg/mL). For this purpose, 1 mL of the compound in the first tube with a concentration of 2 mg/mL was diluted with 1 mL of the MHB culture medium in the second tube so that 1 mL was removed from the first tube and added to the second tube containing 1 mL. This process was repeated up to tube number 9, and then 1 mL was removed from the last tube and ejected, eventually resulting in half dilution of the previous tube. Then, 50 μL of microbial suspension containing 1.5 × 108 bacteria were transferred to the tubes. All the test tubes were incubated for 24 hours at 37 ̊C. After incubation, the tubes were tested for turbidity because of the bacterial growth. All tubes with no bacterial growth were sampled and cultured to determine the minimum bactericidal concentration (MBC) of the compounds. To this end, the tubes showing no bacterial growth were cultured on the MHA culture medium, and the cultured plates were controlled for microbial growth after incubation for 24 hours. The lowest concentration of compounds in the relevant plates, exhibiting bacterial growth failure, was considered as the MBC of that compound (15).
Molecular Docking
Protein Preparation
The crystallographic shape of the Gbp-C of S. mutans with a decision of 1.83°A was changed into a downloaded form from the protein database (PDB ID: 6CAM). The main protein was changed into an organized form for the docking process by casting off all ligand molecules, waters, and extra molecules in addition to the chains of protein not required for docking after that, including polar hydrogen.
Ligand Preparation
As shown in Table 1, the eight derivatives of 1, 3, 4-oxadiazole (4a-h) which are referred to as ligands, were drawn with ChemDraw professional (19.1) software program. In the next step, the ligands were optimized by Chem3D professional (19.1) software program by Molecular mechanics (MM+) and Semi empirical (AM1) methods.
Table 1.
The Structure of 1, 3, 4-oxadiazole Derivatives
4a: (5-(Phenyl)-O)(P)M; 4b: (5-(3-(BromoPhenyl))-O)(P)M; 4c: (5-(3-(ChloroPhenyl))-O)(P)M; 4d: (5-((Naphthalene)-2-il)- O)(P)M; 4e: (5-(3-(FluoroPhenyl))-O)(P)M; 4f: (5-(4-(MethoxyPhenyl))-O)(P)M; 4g: (5-(3-(MethoxyPhenyl))-O)(P)M; 4h: (5-(3, 4-(DiMethoxyPhenyl))-O)(P)M; Note. O: 1, 3, 4-Oxadiazole-2-il; P: Pyridine-2-il; M: Methanol.
Source. SarveAhrabi et al (15).
Docking
Using the AutoDockTools (1.5.6) program, all polar hydrogen atoms were introduced to the protein structure, and the partial masses of the ligand were calculated and introduced with Discovery Studio 4.5 Client program. In addition the Grid box was 48×48×48 dimensional, and dimensions were additionally taken into consideration as X-center = 247.837, Y-center = -23.737, and Z-center = 9.269. Next, the great layout with the least quantity of connection power was decided on via AutoDockVina (16) because the strong layout and the great aggregate in the phrases of (-ΔG bind) became tested using Discovery Studio software (2D&3D) according to previous research (17).
Results
In Vitro
This study focused on evaluating the antibacterial activity of the prepared 1, 3, 4-oxadiazol derivatives (4a-h) moieties in their structures (Table 2). The results of the inhibition zone values for synthesized compounds against S. mutans are illustrated in Figure 1. The obtained data offering an inhibition zone of compounds toward goal bacterium, systems containing naphthalene (4d), fluorophenyl (4e), and dimethoxyphenyl (4h) confirmed the high-quality performance (Table 2). According to Table 2 and Figure 1, evidently, compound 4e, which has a fragrant hydrocarbon and is extensively used for disinfection and insecticide, has a completely favorable impact on the microorganism in conjunction with the oxadiazole’s ring. Compound 4h has one of the fragrant compounds and by the use of pairing the 2 fragrant compounds of methoxyphenyl, has created a good asset toward the studied microorganism.
Table 2.
Chem3D, AutoDockVina, and Antibacterial Results of Compounds (4a-h) by Agar Well Diffusion, MIC, and MBC method (0.5 µg/mL)
|
Compounds
|
Chem3D
|
AutoDockVina
|
Streptococcus mutans
ATCC35668
|
|
Total Energy (kcal/mol)
|
Affinity (kcal/mol) (-∆G
bind
)
|
IZ (mm)
|
MIC (µg/mL)
|
MBC (µg/mL)
|
| 4a |
12.593 |
-8.0 |
12 ± 1.5 |
≤1000 |
1000 |
| 4b |
13.117 |
-7.6 |
10 ± 0.5 |
≤1000 |
1000 |
| 4c |
12.957 |
-7.8 |
16 ± 0.5 |
≤500 |
≤1000 |
| 4d* |
8.732
|
-9
|
19 ± 0.5
|
≤250
|
≤500
|
| 4e |
12.483 |
-8.1 |
20 ± 0.5 |
≤250 |
≤500 |
| 4f |
334.995 |
-7.8 |
16 ± 0.5 |
≤500 |
≤1000 |
| 4g |
137.387 |
-8.4 |
12 ± 1.5 |
≤1000 |
1000 |
| 4h* |
137.286
|
-8.7
|
18 ± 0.5
|
≤250
|
≤500
|
| V |
- |
- |
30±0.5 |
≤125 |
≤250 |
Note. IZ: Inhibition zone; MIC: Minimum inhibitory concentration; MBC: Minimum bactericidal concentration; V: Vancomycin; ± : Averaged three times; *: The best affinity of derivatives to use for docking.
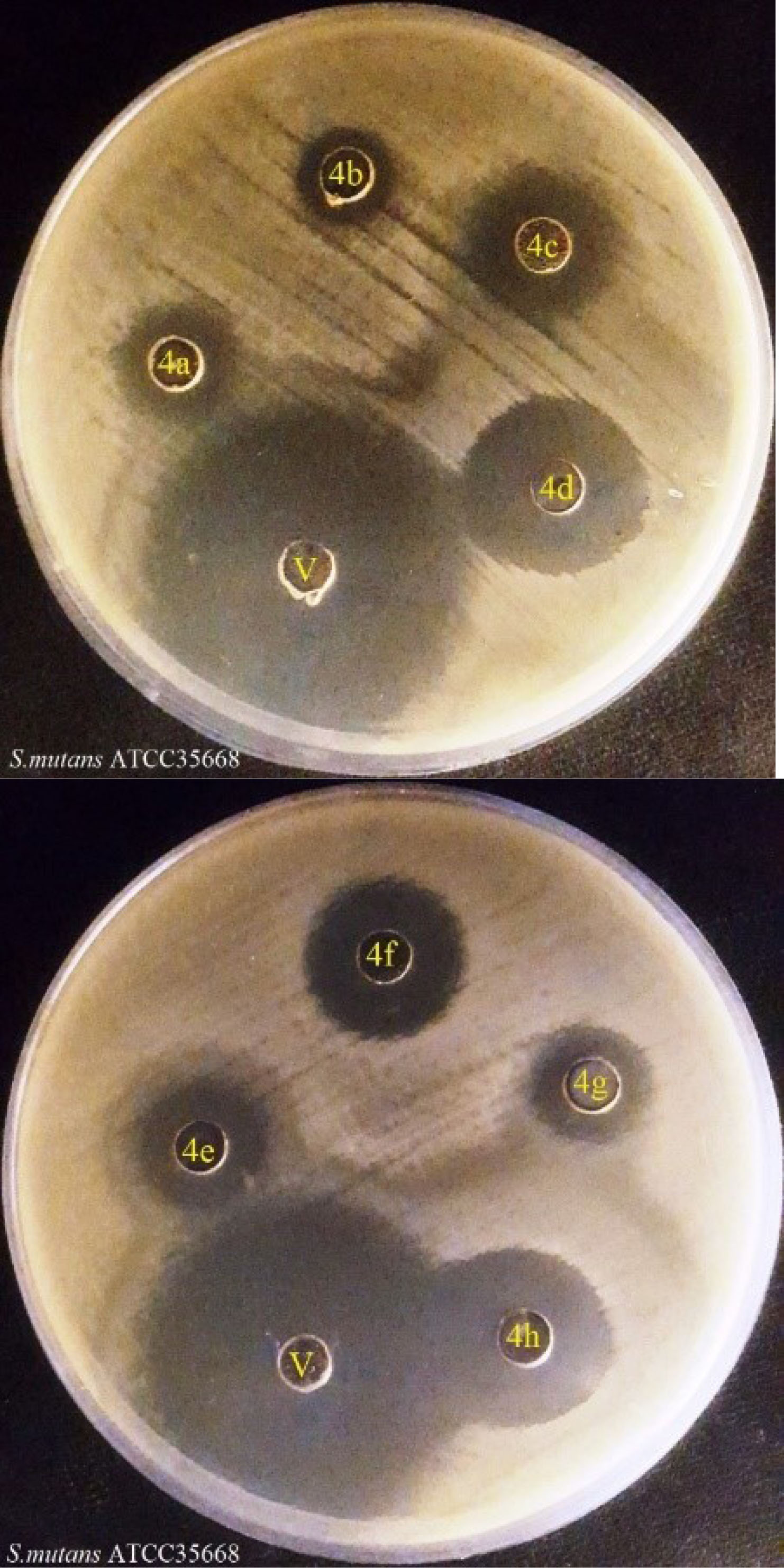
Figure 1.
The Inhibition Zone of Compounds Against S. mutans at 0.5 mg/mL Concentration. Note. S. mutans: Streptococcus mutans; V: Vancomycin.
.
The Inhibition Zone of Compounds Against S. mutans at 0.5 mg/mL Concentration. Note. S. mutans: Streptococcus mutans; V: Vancomycin.
As shown in Table 2, compounds 4h, 4d, and 4e had the highest effect between all compounds (IZ = 18 ± 0.5 mm, MIC = ≥250 µg/mL, MBC = ≥500 µg/mL, IZ = 19 ± 0.5 mm, MIC = ≥250 µg/mL, MBC = ≥500 µg/mL, and IZ = 20 ± 0.5 mm, MIC = ≥250 µg/mL, MBC = ≥500 µg/mL), respectively. These outcomes may be because of the presence of naphthalene (4d), fluorophenyl (4e), dimethoxyphenyl (4h) organizations within the primary compound.
In silico
As shown in Table 2, all the affinities of the compounds were calculated, and high-quality compounds with low ΔG (-ΔG bind) were decided on to hold experiments and inspect chemical interactions (4d and 4h). According to Figure 2, the best quantity of Hydrogen Bonds (HB) as a good way to inhibit the active site of Gbp-C was associated to compound 4h. the compound 4h, by creating hydrogen bonds with amino acid tryptophan (451 HB), threonine (449 HB), glutamate (360 HB), and serine (347 HB) can appear as a powerful compound in inhibiting Gbp-C protein. The opinion that the 4h compound can inhibit Gbp-C is corroborated through the HB formed with tryptophan, isoleucine, threonine and threonine. About 4d, the best quantity of HB belonged to aspartate (402 HB), glutamine (391 HB), glutamate (360 HB), and tryptophan (351 HB). The opinion that the 4d compound can inhibit Gbp-C is corroborated by the HB formed with aspartate and glutamine.
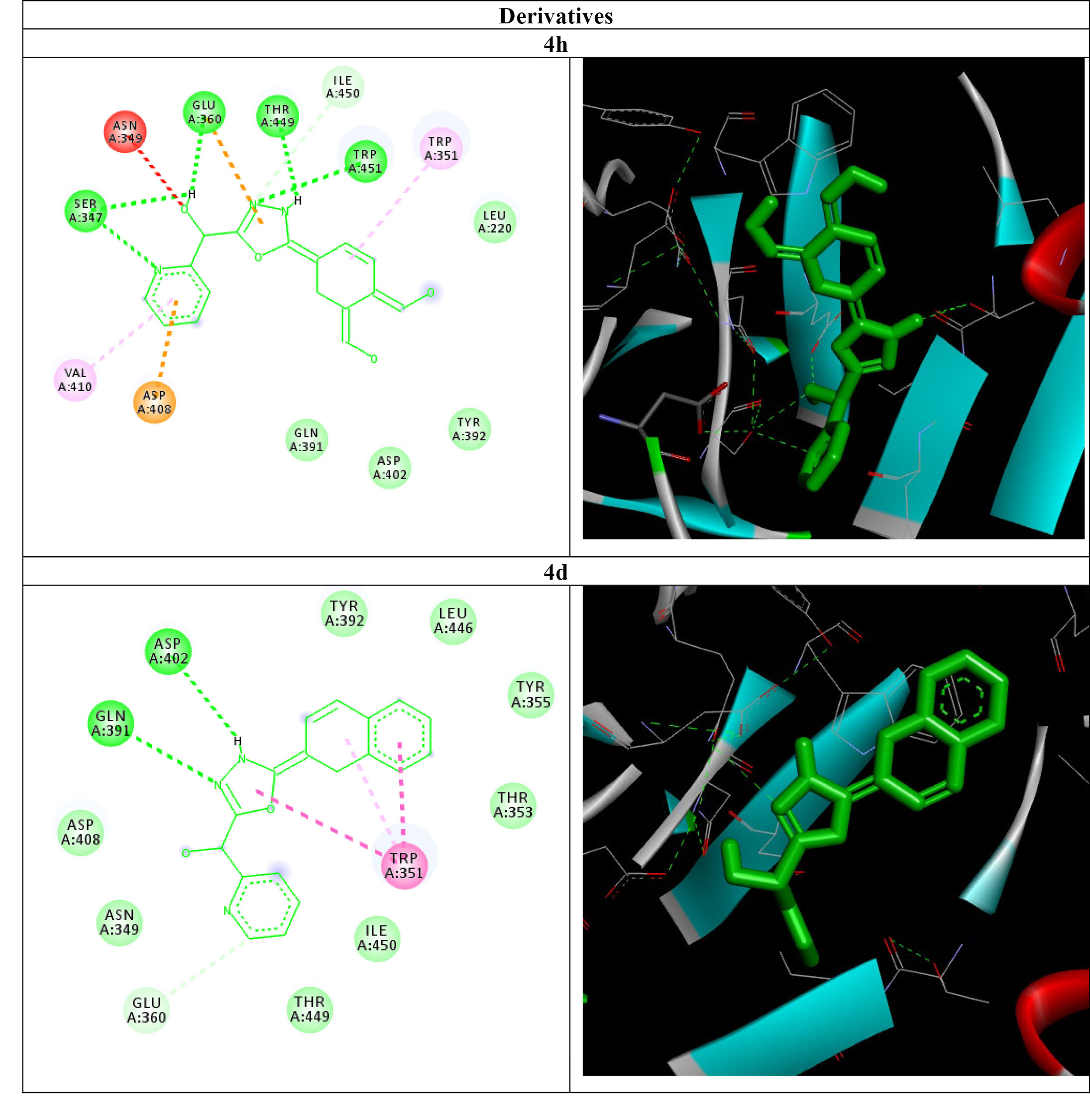
Figure 2.
AutoDockVina Results of Compound 4d and 4h in the Binding site of Gbp-C. Note. Gbp-C: Glucan binding protein-C. Bonds are shown by means of Discovery Studio software (2D&3D).
.
AutoDockVina Results of Compound 4d and 4h in the Binding site of Gbp-C. Note. Gbp-C: Glucan binding protein-C. Bonds are shown by means of Discovery Studio software (2D&3D).
Discussion
Due to the increase in microbial resistance to common drug compounds and the basic need for alternative drug structures in the treatment of diseases, especially microbes related to the mouth and teeth, the synthesis of drug structures is increasing. New structures based on 1, 3, 4-oxadiazole have shown good and biologically acceptable activity. Therefore, their newly synthesized derivatives can be a therapeutic aid for microbial resistance. The present study was performed in vitro and in silico with the aim of the inhibitory effect of 1, 3, 4-oxadiazole derivatives against S. mutans and Gbp-C of S. mutans. In general, 1, 3, 4-oxadiazoles are multi-functional compounds with a diversity of biological properties (e.g., antimicrobial properties and the like), thus their new compound synthesis is important. S. mutans belongs to the streptococci group of mutants and is associated with human caries. More precisely, it is one of the main factors involved in oral diseases. This bacterium affects the tooth \bacteria. One of the limitations of this study is the absence of S. mutans samples isolated from clinical samples and the study of derivatives synthesized on them. Therefore, it is suggested that a synthesized sample be performed on clinical samples as well.
Conclusion
In general, compound 4d with the naphthalene group and 4h with the dimethoxy phenyl group were immobilized to be active against S. mutans. Thus, 4d and 4h as a new series of 1, 3, 4-oxadiazole derivatives can be used for pharmaceutical purposes due to their antibacterial activity and availability, especially in the treatment and replacement of resistance drugs. Finally, it is suggested that future studies investigate bacterial strains isolated from patients in addition to standard strains.
Authors’ Contribution
YA and BO developed the concept and design of the study. YA and BO performed laboratory steps. YA was primarily responsible for writing the manuscript. All authors read and approved the final version.
Ethical Statement
The disclaimer implies that ethical principles have been considered in relation to the proposed work, and no ethical issues were found to be applied to this research proposal.
Conflict of Interest Disclosures
The authors declare that they have no conflict of interests.
Funding
Part of the costs of this project was supported by the Islamic Azad University of Central Tehran Branch.
Acknowledgement
This study was supported by the Department of Biology, Central Tehran Branch, Islamic Azad University, Tehran, Iran.
References
- Peres MA, Macpherson LMD, Weyant RJ, Daly B, Venturelli R, Mathur MR. Oral diseases: a global public health challenge. Lancet 2019; 394(10194):249-60. doi: 10.1016/s0140-6736(19)31146-8 [Crossref] [ Google Scholar]
- Banas JA, Drake DR. Are the mutans streptococci still considered relevant to understanding the microbial etiology of dental caries?. BMC Oral Health 2018; 18(1):129. doi: 10.1186/s12903-018-0595-2 [Crossref] [ Google Scholar]
- Mosaddad SA, Tahmasebi E, Yazdanian A, Rezvani MB, Seifalian A, Yazdanian M. Oral microbial biofilms: an update. Eur J Clin Microbiol Infect Dis 2019; 38(11):2005-19. doi: 10.1007/s10096-019-03641-9 [Crossref] [ Google Scholar]
- Samaranayake L. Essential Microbiology for Dentistry-E-Book. Elsevier; 2018.
- Wilson M, Wilson PJK. Tooth decay. In: Close Encounters of the Microbial Kind: Everything You Need to Know About Common Infections. Cham: Springer; 2021. p. 273-91. 10.1007/978-3-030-56978-5_20.
- Lee SW, Phillips KS, Gu H, Kazemzadeh-Narbat M, Ren D. How microbes read the map: effects of implant topography on bacterial adhesion and biofilm formation. Biomaterials 2021; 268:120595. doi: 10.1016/j.biomaterials.2020.120595 [Crossref] [ Google Scholar]
- Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol 2018; 26(3):229-42. doi: 10.1016/j.tim.2017.09.008 [Crossref] [ Google Scholar]
- Al-Mazini MA. Comparison of biofilm gene (gtf-B) and antibiotics resistance of Streptococcus mutans from dental caries in diabetic and non-diabetic patients. Drug Invent Today 2020; 13(5):802-7. [ Google Scholar]
- De Vuyst L, Vaningelgem F. Developing new polysaccharides. In: McKenna BM, ed. Texture in Food. Woodhead Publishing; 2003. p. 275-320.
- Zarrabi Ahrabi N, Souldozi A, SarveAhrabi Y. In vitro evaluation of antibacterial and antifungal properties of some new 1,3,4-oxadiazole derivatives containing phenyl group. Infect Epidemiol Microbiol 2020; 6(3):177-92. [ Google Scholar]
- SarveAhrabi Y, Zarrabi Ahrabi N, Souldozi A. Synthesis and antimicrobial evaluation of new series of 1,3,4-oxadiazole containing cinnamic acid derivatives. Avicenna J Clin Microbiol Infect 2021; 8(1):11-6. doi: 10.34172/ajcmi.2021.03 [Crossref] [ Google Scholar]
- Li Z, Zhan P, Liu X. 1,3,4-oxadiazole: a privileged structure in antiviral agents. Mini Rev Med Chem 2011; 11(13):1130-42. doi: 10.2174/138955711797655407 [Crossref] [ Google Scholar]
- Aljamali NM, Mahmood RM, Atiya RN, Wannas FA. Review on preparation methods & application fields of oxadiazole. International Journal of Chemical Synthesis and Chemical Reactions 2020; 6(1):49-60. [ Google Scholar]
- Bhati S, Kumar V, Singh S, Singh J. Synthesis, characterization, antimicrobial, anti-tubercular, antioxidant activities and docking simulations of derivatives of 2-(pyridin-3-yl)-1H-benzo[d]imidazole and 1,3,4-oxadiazole analogy. Lett Drug Des Discov 2020; 17(8):1047-59. doi: 10.2174/1570180816666191122105313 [Crossref] [ Google Scholar]
- SarveAhrabi Y, Souldozi A, Zarrabi Ahrabi N. In vitro evaluation of antimicrobial properties of some new 1,3,4-oxadiazole derivatives against Acinetobacter baumannii. Infect Epidemiol Microbiol 2020; 6(1):37-49. doi: 10.29252/iem.6.1.37 [Crossref] [ Google Scholar]
- Barbuceanu SF, Bancescu G, Cretu OD, Draghici C, Bancescu A, Radu-Popescu M. New heterocyclic compounds from 1,3,4-thiadiazole, 1,3,4-oxadiazole and 1,2,4-triazole class with potential antibacterial activity. Rev Chim (Bucharest) 2010; 61(2):140-5. [ Google Scholar]
- Morris GM, Lim-Wilby M. Molecular docking. In: Kukol A, ed. Molecular Modeling of Proteins. Totowa, NJ: Humana Press; 2008. p. 365-82. 10.1007/978-1-59745-177-2_19.