Avicenna J Dent Res. 16(3):146-154.
doi: 10.34172/ajdr.1769
Original Article
A Novel Approach for Apical Periodontitis Prevention Through RIPK3 Inhibition Using Organic Flavonoids
Hamed Karkehabadi 1  , Soroush Sadr 1
, Soroush Sadr 1  , Elham Khoshbin 1
, Elham Khoshbin 1  , Amir Taherkhani 2, *
, Amir Taherkhani 2, * 
Author information:
1Department of Endodontics, School of Dentistry, Hamadan University of Medical Sciences, Hamadan, Iran
2Research Center for Molecular Medicine, Hamadan University of Medical Sciences, Hamadan, Iran
Abstract
Background: Apical periodontitis (AP) entails inflammatory bone resorption facilitated by the induction of necroptosis through receptor-interacting protein kinase 3 (RIPK3). Consequently, the inhibition of RIPK3 holds the potential for favorable effects in addressing AP. A comprehensive investigation, encompassing 44 flavonoids, was undertaken to identify potential inhibitors of RIPK3.
Methods: The crystal structure of the RIPK3 catalytic domain (PDB ID: 7MON) underwent meticulous preparation through the removal of ligands and energy optimization. Overall, 44 flavonoids were examined in this study. Utilizing AutoDock 4.0, molecular docking analyses were performed to evaluate the binding energies within the active site of RIPK3 concerning the examined herbal isolates, with subsequent comparison to the results obtained from a positive control inhibitor. Furthermore, the BIOVIA Discovery Studio Visualizer played a crucial role in elucidating the interactions between the top-ranked flavonoids and the residues of RIPK3.
Results: Five distinct flavonoids—orientin, rutin, vicenin-2, amentoflavone, and nicotiflorin—manifested notable inhibitory effects on RIPK3, boasting inhibition constant values at either the femtomolar or picomolar scale. Notably, the first three members of this set showcased superior binding affinity to the active site of RIPK3 compared to the reference compound.
Conclusion: The proposition arises that flavonoid, particularly orientin, rutin, vicenin-2, amentoflavone, and nicotiflorin, stand as promising herbal isolates for the therapeutic management of AP.
Keywords: Apical periodontitis, Drug, Flavonoid, Inflammations, Molecular docking, Receptor-interacting protein 3
Copyright and License Information
© 2024 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Karkehabadi H, Sadr S, Khoshbin E, Taherkhani A. A novel approach for apical periodontitis prevention through RIPK3 inhibition using organic flavonoids. Avicenna J Dent Res. 2024; 16(3):146-154. doi:10.34172/ajdr.1769
Background
Apical periodontitis (AP) is a prevalent oral inflammatory ailment arising from microbial intrusion into the dental pulp, instigating an inflammatory response and consequential bone deterioration in periapical tissues (1). This infectious disorder, marked by a substantial global prevalence, constitutes a public health concern (2). The research underscores the role of bacterial infection in the periapical region, particularly by Fusobacterium nucleatum, as a pivotal contributor to apical inflammation owing to its potent proinflammatory attributes (3). The distinctive dental composition of the mandible and maxilla facilitates direct transmission of infection to the bone marrow in cases of pulp necrosis, emphasizing the critical role of host immune and tissue responses in constraining the disease (4).
Noteworthy is the fact that approximately 50% of the global adult population exhibits at least one tooth affected by AP, with an even higher prevalence in dental clinic samples (5). These findings underscore the imperative for enhanced preventative measures against this widespread oral disease. Studies reveal that microbial antigens from root canal infections can prompt specific and nonspecific immune responses in periapical tissues. However, factors contributing to the persistence of chronic periapical lesions despite host defense mechanisms remain incompletely elucidated (6).
Recent evidence has implicated necroptosis, a lytic and proinflammatory form of regulated cell death, in the pathogenesis of AP. The formation of the necrosome complex, orchestrated by receptor-interacting protein kinase 3 (RIPK3) and RIPK1, culminates in the phosphorylation of mixed lineage kinase domain-like protein (MLKL), thereby executing necroptosis (7). The activation of the RIPK3-MLKL pathway is meticulously regulated and controlled by microbial and other signaling mechanisms (8). The inhibition of RIPK3 has demonstrated efficacy in diminishing bone destruction and inflammation in AP models, thereby accentuating its potential as a therapeutic target (9).
Flavonoids represent a substantial category of polyphenolic compounds widely distributed in fruits, vegetables, grains, herbs, and various plant sources. Their diverse bioactivities, including anti-inflammatory, anticancer, antioxidant, and antimicrobial properties, have undergone extensive investigation (10). Recent findings underscore the capacity of flavonoids to modulate pivotal proteins associated with inflammation, cell survival, proliferation, and other essential cellular processes (11).
Molecular docking, a computational method, is valuable in identifying potential inhibitors against therapeutic targets by assessing their binding affinity to active sites and allosteric pockets (12). Compared to conventional experimental techniques, in silico screening offers a rapid and cost-effective approach to drug discovery (13). In the present study, molecular docking is employed to scrutinize a library of flavonoids, aiming to ascertain their potential as inhibitors of RIPK3. It is aimed to investigate these plant-derived compounds’ binding modes and interactions with the catalytic domain of RIPK3. Compounds exhibiting favorable binding energy scores are identified as hits, demonstrating potential as RIPK3 inhibitors. These flavonoid hits emerge as promising drug candidates, warranting further experimental validation in subsequent studies for their development as novel therapeutics against inflammatory conditions, such as AP. Utilizing in silico methods facilitates the efficient prioritization of compounds with optimally predicted bioactivity. The overview of the present study design is presented in Figure 1.
Figure 1.
A Schematic Illustration of the Present Study
.
A Schematic Illustration of the Present Study
Materials and Methods
RIPK3 Structure Preparation
The crystallographic data of the catalytic domain of RIPK3 were sourced from the Research Collaboratory for Structural Bioinformatics under the accession code 7MON, achieving a resolution of 2.24 Å (14,15). Within the obtained Protein Data Bank (PDB) file, two distinct polypeptide chains were identified, including chain A corresponding to the mixed lineage kinase domain-like protein and chain B representing the 316-residue RIPK3 kinase domain selected for subsequent molecular docking investigations. Preceding the docking procedures, the exclusion of non-essential heteroatoms, encompassing water molecules, metals, and the co-crystallized ligand compound N-[4-({2-[(cyclopropanecarbonyl)amino]pyridin-4-yl}oxy)-3-fluorophenyl]-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide ([ZL1], a positive control inhibitor with PubChem ID 155524868), was executed on the RIPK3 PDB file using the molecular visualization software Notepad + + . The binding interactions between ZL1 and crucial amino acid residues within the active site of RIPK3 were assessed through visual analysis utilizing the BIOVIA Discovery Studio Visualizer, version 19.1.0.18287. This examination aimed to discern key interacting residues, thereby facilitating the specification of the binding site for subsequent investigations involving flavonoid ligands.
After the retrieval, the acquired RIPK3 structure underwent a process of energetic minimization to attain its utmost stable conformation. This optimization procedure employed the Swiss-PdbViewer software (version 4.0.1), wherein emphasis was placed on refining the hydrogen bonding network and alleviating steric clashes. This optimization entailed iterative steps involving steepest descent minimization, followed by conjugated gradient minimization, persisting until convergence. The resulting optimized structure, characterized by the lowest energy state and the most favorable geometric coordinates, was then preserved in PDB format, facilitating its utilization in subsequent docking analyses.
Ligands Structure Preparation
The three-dimensional (3D) configurations of 44 natural flavonoids (16,17) were sourced from the National Center for Biotechnology Information PubChem compound database in a structured data format (SDF) file. Subsequently, these structures underwent conversion to the PDB format utilizing the online SMILES Translator and Structure File Generator tool known as Cactus, accessible at https://cactus.nci.nih.gov/translate/.
Then, the energetic minimization of ligands was executed using the molecular modeling software HyperChem (version 8.0.10) to attain their most stable conformations. Employing the Molecular Mechanics (MM + ) force field, structural refinement was achieved through 500 steps of steepest descent minimization, followed by convergence utilizing the Polak–Ribière-Polyak conjugate gradient algorithm. The resulting optimized structures of flavonoids, characterized by their lowest energy conformations, were then systematically saved in PDB format, serving as the basis for subsequent docking experiments. In evaluating the binding affinity of flavonoids, a comparative analysis was conducted with ZL1, employed as a positive control inhibitor (18).
Binding Site Residues
The binding site of the RIPK3 kinase domain comprised crucial interacting residues determined from an earlier structural analysis of the co-crystallized reference inhibitor complex (18). These residues, namely, Val27, Val35, Ala48, Lys50, Val52, Glu60, Met64, Leu92, Kys95, Phe96, Met97, Leu149, Ala159, Asp160, and Phe161, play significant roles in forming essential hydrogen bonds and hydrophobic interactions with both flavonoid and ZL1.
Molecular Docking
Molecular docking analysis was used to assess the binding affinities between the active site of RIPK3 and a curated collection of 44 flavonoids, employing AutoDock 4.0 tools. The research was conducted on a Windows-based Intel Core i5 PC system with 16 GB RAM. AutoDock utilizes a Lamarckian genetic algorithm, iteratively modifying a ligand pose population within the binding site through random alterations, followed by localized optimization, to generate valid docking conformations.
The catalytic cleft of RIPK3 was defined as a 3D grid box with 68 x 64 x 50 points in the x, y, and z axes. The center coordinates were set at x = -41.621, y = 4.48, and z = -19.041 Å, and the grid points had a spacing of 0.375 Å (18). This grid encompassed the crucial interacting residues within the RIPK3 catalytic domain, as identified in a prior analysis of the reference ligand complex. Hydrogen atoms were introduced to the protein, and charges were assigned based on Gasteiger partial charges. For all flavonoids, alongside the reference compound ligands, active torsions were designated for all rotatable bonds, and non-polar hydrogens were amalgamated.
Each ligand underwent 50 individual docking runs, yielding a single docked pose per run. The genetic algorithm generated a population of 150 random ligand conformations in each run, with a maximum of 2.5 × 107 energy evaluations and a limit of 27 000 generations. The docking results were clustered using a root mean square deviation tolerance of 2.0 Å. The docked pose exhibiting the most favorable free energy of binding within the most significant cluster was chosen as the optimal binding mode for subsequent analysis.
The approximation of binding affinity between each flavonoid ligand and the RIPK3 protein relied on the docking score, determined as the change in the estimated free energy of binding (ΔGbinding) measured in kcal/mol units. Lower ΔGbinding values were indicative of stronger predicted binding affinities. Additionally, the estimated inhibition constant (Ki) for each flavonoid-RIPK3 complex was derived from the docking score. Compounds characterized by more negative ΔGbinding and Ki values were classified as hits, suggesting favorable predicted inhibition of RIPK3. Finally, the exchanges between ligands and the 7MON active site were visually represented using the BIOVIA Discovery Studio Visualizer.
Results
Binding Affinity Assessment
Orientin emerged as the most potent RIPK3 inhibitor based on molecular docking analysis, exhibiting a favorable Ki value at the femtomolar (fM) scale. Additionally, rutin, vicenin-2, amentoflavon, and nicotiflorin were predicted to impede the enzyme’s activity at the picomolar (pM) concentration. Consequently, these five herbal isolates were identified as top-ranked RIPK3 inhibitors among the studied flavonoids. The ΔGbinding values for orientin, rutin, vicenin-2, amentoflavon, and nicotiflorin with RIPK3 were calculated as -16.42, -15.97, -14.55, -13.01, and -12.91 kcal/mol, respectively. Furthermore, the ΔGbinding score and the Ki value for ZL1 were estimated at -13.18 and 219.38 pM, respectively. Orientin, rutin, and vicenin-2 notably exhibited higher binding affinity to the RIPK3 active site than the reference compound, as indicated by the ΔGbinding score (Figure 2). Table 1 provides a comprehensive compilation of ΔGbinding and Ki values for 44 flavonoids concerning the active sites of ZL1 and RIPK3. Meanwhile, Table 2 delineates the energy specifics of the top-ranked flavonoids and their interaction with the RIPK3 enzyme.
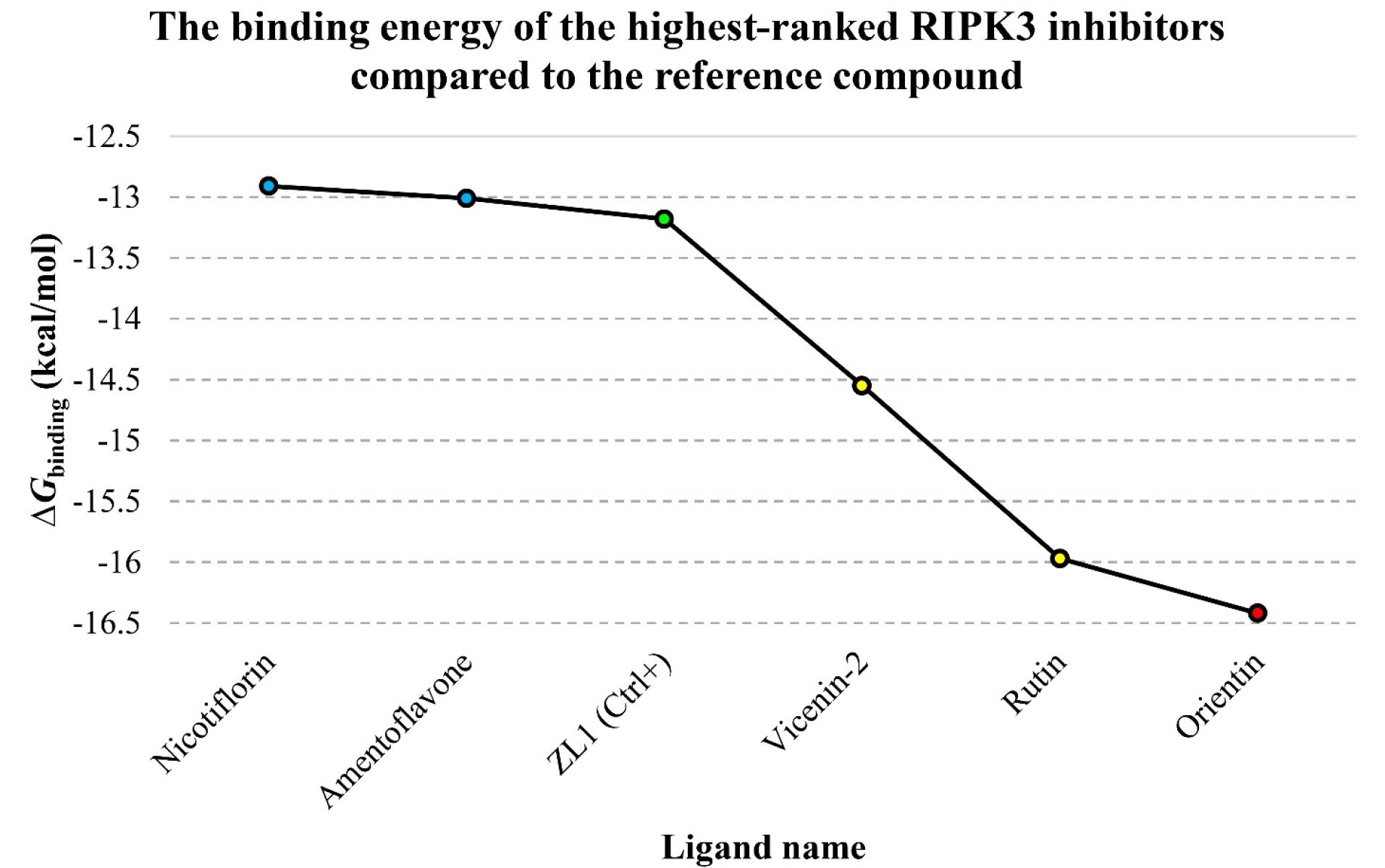
Figure 2.
The Graph of ΔGbinding Values in kcal/mol Units for the Top-Ranked Flavonoids, ZL1, and the RIPK3 Active Site. Note. Ligand names are on the X-axis, while the corresponding Gibbs free binding energy is plotted on the Y-axis. Notably, the green diamond symbolizes ZL1, the red spot denotes the most potent RIPK3 inhibitor, and the yellow and blue spots represent compounds exhibiting higher and lower binding affinities to RIPK3 than the reference inhibitor. RIPK3: Receptor-interacting serine/threonine-protein kinase 3
.
The Graph of ΔGbinding Values in kcal/mol Units for the Top-Ranked Flavonoids, ZL1, and the RIPK3 Active Site. Note. Ligand names are on the X-axis, while the corresponding Gibbs free binding energy is plotted on the Y-axis. Notably, the green diamond symbolizes ZL1, the red spot denotes the most potent RIPK3 inhibitor, and the yellow and blue spots represent compounds exhibiting higher and lower binding affinities to RIPK3 than the reference inhibitor. RIPK3: Receptor-interacting serine/threonine-protein kinase 3
Table 1.
Calculated Gibbs Free Energy of Binding and Ki Values Between the RIPK3 Active Site and 44 Natural Flavonoids
|
Ligand ID
|
Ligand Name
|
Binding Energy (kcal/mol)
|
Ki
|
| 5281675 |
Orientin |
-16.42 |
927.76 fM |
| 5280805 |
Rutin |
-15.97 |
1.96 pM |
| 442664 |
Vicenin-2 |
-14.55 |
21.57 pM |
| 5281600 |
Amentoflavone |
-13.01 |
289.10 pM |
| 5318767 |
Nicotiflorin |
-12.91 |
344.76 pM |
| 72936 |
Sophoraflavanone G |
-12.03 |
1.52 nM |
| 9911508 |
Astragarin |
-11.99 |
1.63 nM |
| 5280441 |
Vitexin |
-11.98 |
1.66 nM |
| 5280804 |
Isoquercitrin |
-11.8 |
2.24 nM |
| 5353915 |
Quercetin-3-rhamnoside |
-11.7 |
2.64 nM |
| 5280459 |
Quercitrin |
-11.67 |
2.80 nM |
| 5280637 |
Cynaroside |
-11.57 |
3.30 nM |
| 10095180 |
Kaempferol 7-O-glucoside |
-11.26 |
5.53 nM |
| 5282102 |
Astragalin |
-10.82 |
11.67 nM |
| 5280343 |
Quercetin |
-10.56 |
18.14 nM |
| 5281612 |
Diosmetin |
-10.1 |
39.76 nM |
| 5280445 |
Luteolin |
-10.02 |
44.85 nM |
| 5280704 |
Apigenin-7-glucoside |
-10.01 |
46.10 nM |
| 439533 |
Taxifolin |
-9.89 |
56.72 nM |
| 5281654 |
Isorhamnetin |
-9.48 |
112.27 nM |
| 5280681 |
3-O-Methylquercetin |
-9.41 |
125.84 nM |
| 5281672 |
Myricetin |
-9.38 |
132.80 nM |
| 471 |
Dihydroquercetin |
-8.98 |
263.73 nM |
| 9064 |
Catechin |
-8.97 |
263.78 nM |
| 5280544 |
Herbacetin |
-8.95 |
275.11 nM |
| 124052 |
Glabridin |
-8.87 |
317.04 nM |
| 5316673 |
Afzelin |
-8.87 |
314.36 nM |
| 629440 |
Hemileiocarpin |
-8.55 |
542.31 nM |
| 5280863 |
Kaempferol |
-8.52 |
568.26 nM |
| 5318998 |
Licochalcone A |
-8.47 |
616.74 nM |
| 10680 |
Flavone |
-8.46 |
627.06 nM |
| 5281614 |
Fisetin |
-8.38 |
725.50 nM |
| 639665 |
XanthohuMol |
-8.34 |
771.84 nM |
| 5281607 |
Chrysin |
-8.32 |
803.49 nM |
| 5280443 |
Apigenin |
-8.18 |
54.78 nM |
| 638278 |
Isoliquiritigenin |
-8.16 |
1.04 uM |
| 14309735 |
Xanthogalenol |
-8.16 |
1.04 uM |
| 5317435 |
Fustin |
-8.15 |
1.06 uM |
| 5281670 |
Morin |
-8.07 |
1.22 uM |
| 72281 |
Hesperetin |
-7.89 |
1.64 uM |
| 1203 |
Epicatechin |
-7.87 |
1.72 uM |
| 5280378 |
Formononetin |
-7.72 |
2.21 uM |
| 25201019 |
Ponciretin |
-7.49 |
3.21 uM |
| 443639 |
Epiafzelechin |
-7.42 |
3.61 uM |
| 155524868 |
ZL1 (Ctrl + ) |
-13.18 |
219.38 pM |
Table 2.
All Types of Binding Energy Between the RIPK3 Active Site and Top-Ranked Flavonoids
|
Top-ranked Flavonoids
|
Intermolecular Energy (kcal/mol)
|
Total Internal Energy (kcal/mol)
|
Torsional Free Energy (kcal/mol)
|
Unbound System’s Energy (kcal/mol)
|
∆GBinding
(kcal/mol)
|
| Orientin |
-9.74 |
-11.14 |
3.58 |
-0.89 |
-16.42 |
| Rutin |
-12.05 |
-12.37 |
5.07 |
-3.38 |
-15.97 |
| Vicenin-2 |
-12.28 |
-9.77 |
5.07 |
-2.42 |
-14.55 |
| Amentoflavone |
-11.07 |
-6.15 |
3.28 |
-0.93 |
-13.01 |
| Nicotiflorin |
-10.05 |
-10.41 |
4.77 |
-2.78 |
-12.91 |
Note. RIPK3:Receptor-interacting serine/threonine-protein kinase 3.
Interaction Modes
The analysis of the binding modes revealed that among the docked ligands, rutin established the highest number of hydrogen bonds (n = 9) with RIPK3 residues. Additionally, Amentoflavone displayed the most excellent count of hydrophobic interactions (n = 11). In comparison, the reference inhibitor ZL1 formed 6 H-bonds and 13 hydrophobic interactions. The specific interactions of top-ranked flavonoids with the RIPK3 active site residues are outlined in Table 3 and Figure 3. Figure 4 illustrates the 3D coordinates of orientation within the RIPK3 catalytic cleft.
Table 3.
Interaction Mods Between the RIPK3 Catalytic Cleft and Top-Ranked Flavonoids
|
Ligand Name
|
Hydrogen Bond (Distance in Å)
|
Hydrophobic Interaction (Distance in Å)
|
| Orientin |
Lys50 (4.60), THR (2.83, 4.29), Met97 (4.13, 4.47), Asp160 (3.94, 3.98) |
Val27 (6.41), Val35 (4.46, 4.52), Ala48 (4.00, 4.48), Leu149 (6.27) |
| Rutin |
Gly33 (2.99), Lys95 (6.57), Met97 (3.28, 4.11, 4.20, 5.82), Ser101 (3.13, 4.15), Asp160 (3.85) |
Val27 (4.33), Val35 (4.69), Lys50 (4.48, 4.87), Leu73 (6.09), Ala159 (5.93) |
| Vicenin-2 |
Met97 (3.85), Gly98 (4.04, 5.08), Ser101 (3.87), Ser146 (3.77), Asp160 (3.48) |
Val35 (5.58, 5.78), Ala48 (5.09, 6.08), Lys50 (5.55), Leu73 (5.69), Leu149 (5.18), Ala159 (6.25) |
| Nicotiflorin |
Gly33 (3.67,3.81,3.90), Ala48 (4.54), Leu92 (4.06), Met97 (3.69, 4.00), Asp160 (5.31) |
Val27 (6.30), Val35 (4.71, 4.84), Ala48 (4.85, 5.36), Lys50 (4.54), Phe96 (5.86), Leu149 (6.07) |
| Amentoflavone |
Asp160 (3.42, 3.64, 4.42), Met97 (4.09, 4.79), Ser146 (4.35) |
Val27 (6.11, 6.71), Val35 (4.88, 4.91), Ala48 (4.26, 7.38), Lys50 (5.83), Phe96 (5.73), Leu149 (6.24, 6.34), Ala159 (6.49) |
Note. RIPK3: Receptor-interacting serine/threonine-protein kinase 3.
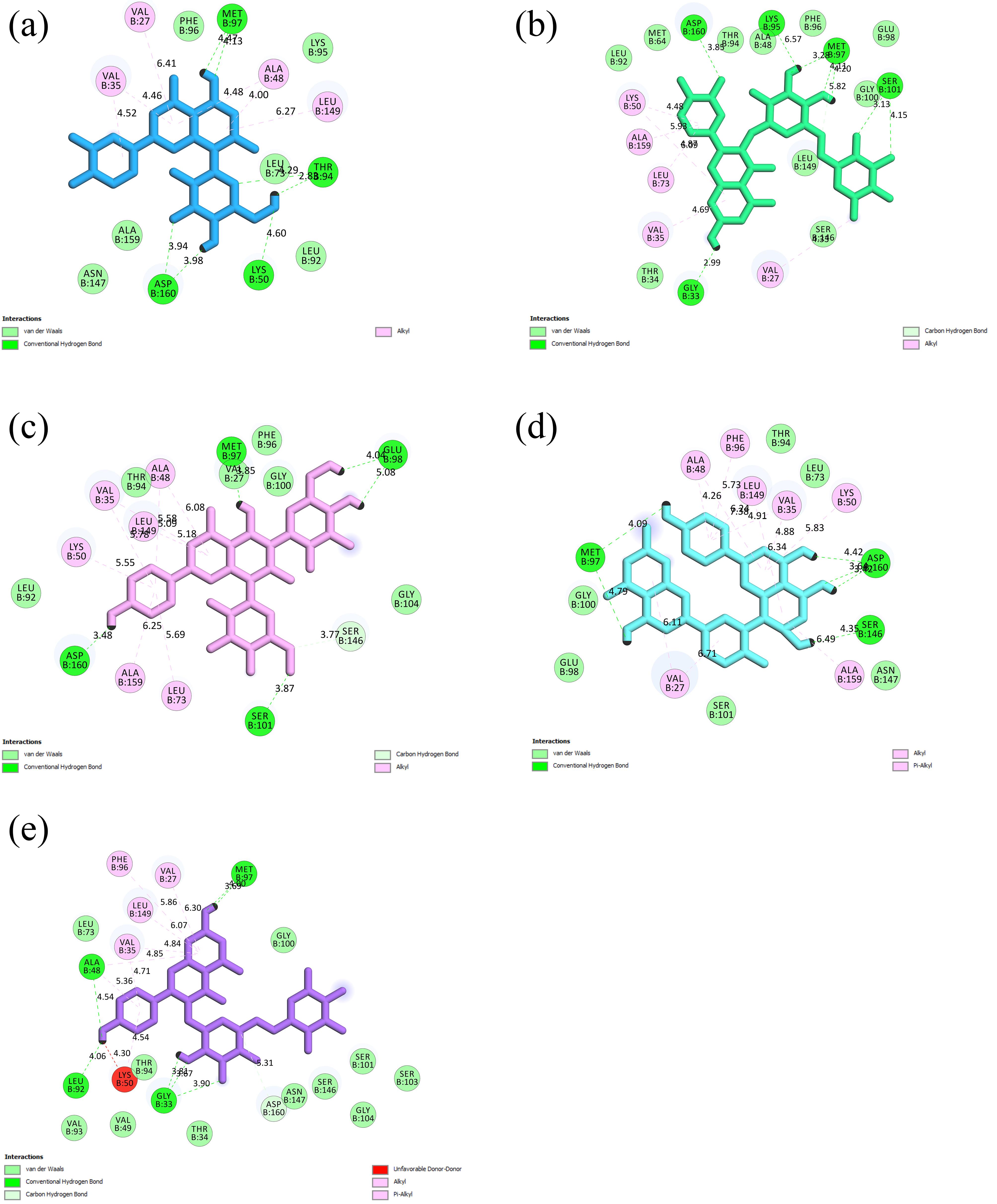
Figure 3.
Interactions Among the Top-Ranked Flavonoids and Residues Within the Active Site of RIPK3 Illustrated for Each Ligand: (a) Orientin, (b) Rutin, (c) Vicenin-2, (d) Amentoflavone, and (e) Nicotiflorin. Note. RIPK3: Receptor-interacting serine/threonine-protein kinase 3
.
Interactions Among the Top-Ranked Flavonoids and Residues Within the Active Site of RIPK3 Illustrated for Each Ligand: (a) Orientin, (b) Rutin, (c) Vicenin-2, (d) Amentoflavone, and (e) Nicotiflorin. Note. RIPK3: Receptor-interacting serine/threonine-protein kinase 3
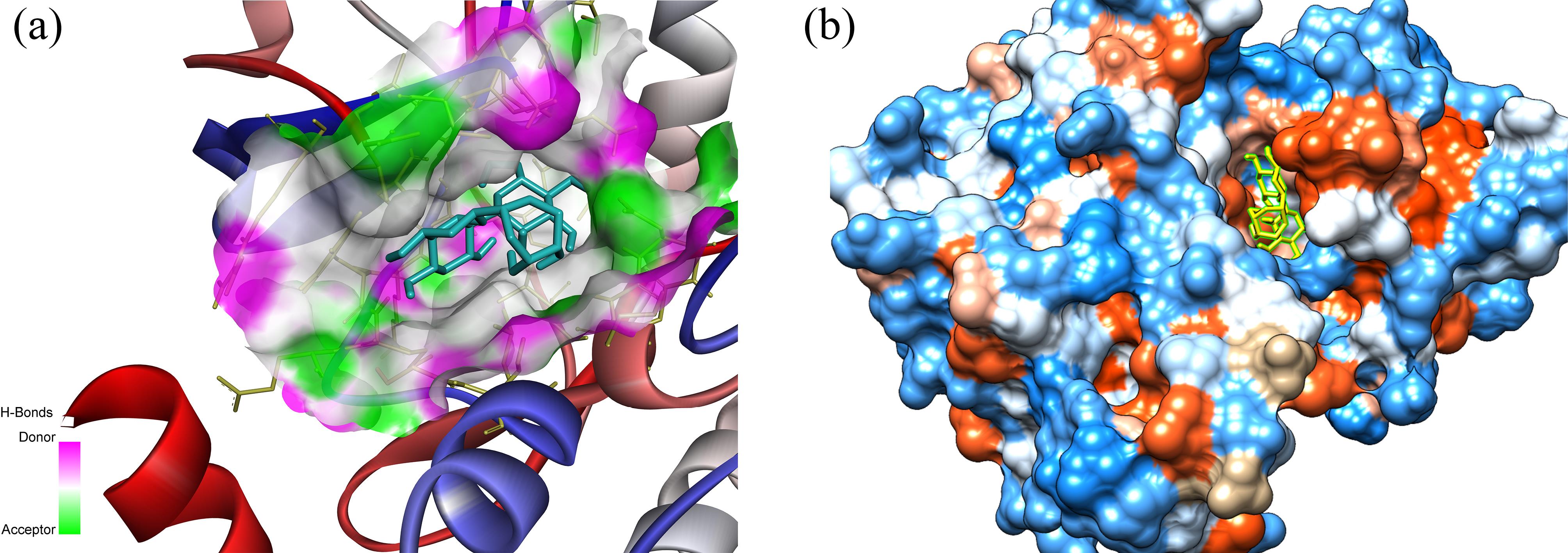
Figure 4.
A Three-dimensional Representation of Orientin Within the Active Site of RIPK3 Depicted in Two Formats, including (a) Ribbon and (b) Hydrophobic Surface Illustrations. Note. RIPK3: Receptor-interacting serine/threonine-protein kinase 3; MD: Molecular dynamics
.
A Three-dimensional Representation of Orientin Within the Active Site of RIPK3 Depicted in Two Formats, including (a) Ribbon and (b) Hydrophobic Surface Illustrations. Note. RIPK3: Receptor-interacting serine/threonine-protein kinase 3; MD: Molecular dynamics
Discussion
AP represents a prevalent oral inflammatory condition with consequential implications for bone integrity and tooth retention (19). Contemporary investigations have associated necroptosis, a proinflammatory manifestation of programmed cell death mediated by RIPK3, with the pathogenesis of AP (1,4). The formation of the necrosome complex involving RIPK1 leads to the phosphorylation of MLKL and instigates lytic cell death (20,21). This necroptotic cascade is presumed to significantly contribute to periapical tissue deterioration and inflammatory bone resorption in the context of AP. Prior research demonstrated that inhibiting RIPK3 can mitigate periapical bone loss in murine models, suggesting its potential as a promising therapeutic target for this widespread oral ailment (1). In addition, the mediation of necroptosis in response to inflammatory factors such as tumor necrosis factor (TNF-α) is facilitated by the interaction between RIPK3 and MLKL proteins. The multifaceted roles of RIPK3 beyond necroptosis, highlighting its contributions to sterile inflammation and tissue damage, have been demonstrated in previous studies. The complexity of RIPK3 signaling pathways and their regulation by other cellular proteins, including caspase-8 and inhibitors of apoptosis protein (IAP) ubiquitin ligases, is underscored. The interaction of RIPK3 with various signaling molecules, downstream of receptors such as the TNF receptor family, interferon-regulatory factors, and toll-like receptors, and its contribution to pro-inflammatory functions has been previously reported. The evidence indicates that RIPK3 acts at the nexus of life and death, influencing the fate of cells and the inflammatory milieu within tissues (22-24). Therefore, it might be suggested that RIPK3 inhibition downregulates several signaling pathways linked to the TNF receptor family, interferon-regulatory factors, and toll-like receptors. Consequently, the current investigation employed a computational drug discovery method to identify prospective RIPK3 inhibitors derived from natural flavonoids.
Within the scope of this study, molecular docking analysis was conducted involving 44 distinct flavonoids against the catalytic domain of RIPK3 to discern potential RIPK3 inhibitors. orientin, rutin, and vicenin-2 emerged as standout compounds, attaining top rankings. Projections based on docking scores suggest that these compounds exhibit a more robust potential for RIPK3 inhibition than the reference inhibitor ZL1, signifying their enhanced affinity for the catalytic domain.
Orientin has demonstrated a notably robust binding affinity to the active site of RIPK3, substantiated by a ∆Gbinding score of -16.42 kcal/mol and a Ki value of 927.76 fM. This flavonoid is characterized by a glucosyl moiety attached to the luteolin aglycone structure. The interaction profile reveals seven hydrogen bonds and six hydrophobic interactions involving Val27, Val35, Ala48, Lys50, Thr94, Met97, Leu149, and Asp160. Prior investigations have highlighted its antioxidative, anti-inflammatory, and antimicrobial properties (25). Abundantly found in passionfruit, basil, and various edible berries (26), orientin has garnered attention for its capacity to inhibit migration, proliferation, and metastasis-associated factors, including metalloproteinases in cancer cells, suggesting therapeutic potential (27,28). The predicted high-affinity RIPK3 inhibition suggests a potential role in suppressing necroptosis, underscoring orientin’s promise as a multifunctional flavonoid warranting further exploration as a natural product targeted toward RIPK3.
In a study by Xiao et al (29), orientin’s potential anti-inflammatory and antioxidant effects were investigated in a lipopolysaccharide-induced acute lung injury (ALI) mouse model [1]. Orientin effectively alleviated lipopolysaccharide-induced ALI, improving lung histology, reducing the W/D ratio, and lowering protein levels in bronchoalveolar lavage fluid. It inhibited key inflammatory mediators (inducible nitric oxide synthase, cyclooxygenase-2, and high mobility group box 1) and demonstrated antioxidant properties by reducing malondialdehyde and reactive oxygen species while enhancing glutathione and superoxide dismutase levels. Orientin also suppressed NLRP3 inflammasome activation and the nuclear factor kappa B signaling pathway, crucial regulators of inflammation. The study highlighted orientin’s impact on restoring nuclear factor erythroid 2-related factor 2 expression and its downstream antioxidant genes. These findings suggest orientin’s potent anti-inflammatory and antioxidant effects, indicating its potential protective role against ALI through NLRP3, nuclear factor kappa B modulation, and nuclear factor erythroid 2-related factor 2 pathway upregulation.
Rutin, characterized by rutinose and glucose bound to quercetin, emerged as another highly-ranked flavonoid inhibitor in this study. The calculated ∆Gbinding and Ki values for the interaction between rutin and the RIPK3 active site were determined to be -15.97 kcal/mol and 1.98 pM, respectively. Rutin exhibited a noteworthy interaction profile, engaging in nine hydrogen bonds and six hydrophobic interactions with Val27, Gly33, Val35, Lys50, Leu73, Lys95, Met97, Ser101, Ala159, and Asp160.
Rutin has garnered attention for its potential anti-inflammatory effects. In wound healing, Saha et al (30) investigated rutin nanoparticles (RNPs), showcasing their anti-inflammatory and antioxidant properties in vitro. RNPs, incorporated into a bilayered skin substitute, demonstrated increased interleukin-10 levels, controlled reactive oxygen species, and reduced fibrosis marker α-smooth muscle actin in fibroblasts. In vivo studies with RNP-integrated substitutes represented enhanced wound healing and re-epithelialization. Hamad et al (31) also explored rutin’s anti-inflammatory and immunomodulatory effects in a murine Schistosoma mansoni infection model, revealing a significant impact on liver pathology, egg entrapment, and proinflammatory cytokine levels. These findings underscore rutin’s robust anti-inflammatory properties, aligning with our study results.
The identified flavonoids, including orientin and rutin, exhibit promise as potential candidates for the development of RIPK3-targeted inhibitors for the treatment of AP. Their predicted ability to bind RIPK3 implies the potential to suppress necroptotic cell death and mitigate inflammatory bone resorption induced by the microbial infection of the dental pulp. However, it is imperative to emphasize that experimental validation through cell-based assays and animal models is essential to confirm these compounds’ bioactivity and protective effects against periapical tissue damage.
While the present study identified promising flavonoid candidates, namely, orientin, rutin, vicenin-2, amentoflavone, and nicotiflorin, as potential inhibitors of RIPK3 for the therapeutic management of AP, further research and optimization efforts are warranted to translate these findings into clinical applications. Structural modifications of the identified flavonoids could be explored to enhance their binding affinity, specificity, or pharmacokinetic properties. Rational drug design approaches, structure-activity relationship studies, and computer-aided drug design techniques can guide the systematic modification of these compounds’ chemical structures, potentially leading to improved inhibitory potency or desirable pharmacological profiles. Formulation strategies should also address potential challenges in delivering flavonoids, such as poor solubility, stability, or bioavailability. Nanoparticle encapsulation, prodrug design, or co-administration with absorption enhancers could be explored to improve the delivery and bioavailability of the identified flavonoid candidates. Furthermore, combination therapies involving the identified flavonoids and existing therapeutic agents for AP or other inflammatory conditions may offer synergistic effects or complementary mechanisms of action, potentially enhancing the overall therapeutic efficacy. Extensive in vitro studies are necessary to validate the inhibitory effects of the identified flavonoids on RIPK3 and related pathways. Additionally, in vivo studies using appropriate animal models should be conducted to evaluate these promising flavonoid candidates’ efficacy, safety, and pharmacokinetic profiles. Translating these findings to clinical settings will require rigorous clinical trials to assess the identified flavonoids’ safety, effectiveness, and tolerability or optimized derivatives in patients with AP or related conditions. Addressing potential challenges in clinical translation, such as scaling up production, establishing appropriate dosing regimens, and monitoring for possible adverse effects, will be crucial for successfully developing these compounds as therapeutic agents.
Conclusion
The ongoing investigation suggests that orientin, rutin, vicenin-2, amentoflavone, and nicotiflorin manifest noteworthy binding affinities toward the active site of RIPK3. These findings hold substantial implications for researchers aiming to pioneer innovative drug therapies for diverse inflammation-related conditions, such as AP. However, it is imperative to underscore that further scrutiny is requisite to authenticate the current observations, encompassing rigorous in vitro and in vivo validation experiments.
Acknowledgments
The authors would like to thank the Research Center for Molecular Medicine, Hamadan University of Medical Sciences, Hamadan, Iran, for their support.
Authors’ Contribution
Conceptualization: Amir Taherkhani, Hamed Karkehabadi.
Data curation: Amir Taherkhani, Soroush Sadr.
Formal analysis: Amir Taherkhani, Soroush Sadr.
Investigation: Amir Taherkhani, Soroush Sadr.
Methodology: Amir Taherkhani, Soroush Sadr.
Project administration: Amir Taherkhani.
Resources: Amir Taherkhani, Hamed Karkehabadi.
Software: Amir Taherkhani, Soroush Sadr.
Supervision: Amir Taherkhani.
Validation: Amir Taherkhani.
Visualization: Amir Taherkhani.
Writing–original draft: Amir Taherkhani.
Writing–review & editing: Hamed Karkehabadi, Elham Khoshbin.
Competing Interests
None declared.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethical Approval
The present study was approved by the Ethics Committee of Hamadan University of Medical Sciences, Hamadan, Iran (Ethical No. IR.UMSHA.REC.1402.449).
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- Liu J, Wang J, Ren J, Yang Q, Zhan W, Wang M. Inhibition of receptor-interacting protein kinase-3 in the necroptosis pathway attenuates inflammatory bone loss in experimental apical periodontitis in Balb/c mice. Int Endod J 2021; 54(9):1538-47. doi: 10.1111/iej.13534 [Crossref] [ Google Scholar]
- Ng YL, Mann V, Gulabivala K. Outcome of secondary root canal treatment: a systematic review of the literature. Int Endod J 2008; 41(12):1026-46. doi: 10.1111/j.1365-2591.2008.01484.x [Crossref] [ Google Scholar]
- Siqueira JF Jr, Rôças IN, Ricucci D, Hülsmann M. Causes and management of post-treatment apical periodontitis. Br Dent J 2014; 216(6):305-12. doi: 10.1038/sj.bdj.2014.200 [Crossref] [ Google Scholar]
- Wang L, Zhang H, Dong M, Zuo M, Liu S, Lu Y. Role of the Btk-PLCγ2 signaling pathway in the bone destruction of apical periodontitis. Mediators Inflamm 2019; 2019:8767529. doi: 10.1155/2019/8767529 [Crossref] [ Google Scholar]
- Taira TM, Lima V, Prado DS, Silva TA, Issa JP, da Silva LA. NLRP12 attenuates inflammatory bone loss in experimental apical periodontitis. J Dent Res 2019; 98(4):476-84. doi: 10.1177/0022034518820289 [Crossref] [ Google Scholar]
- Nikolic N, Jakovljevic A, Carkic J, Beljic-Ivanovic K, Miletic M, Soldatovic I. Notch signaling pathway in apical periodontitis: correlation with bone resorption regulators and proinflammatory cytokines. J Endod 2019; 45(2):123-8. doi: 10.1016/j.joen.2018.10.015 [Crossref] [ Google Scholar]
- Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature 2015; 517(7534):311-20. doi: 10.1038/nature14191 [Crossref] [ Google Scholar]
- Dai X, Deng Z, Liang Y, Chen L, Jiang W, Zhao W. Enterococcus faecalis induces necroptosis in human osteoblastic MG63 cells through the RIPK3 / MLKL signalling pathway. Int Endod J 2020; 53(9):1204-15. doi: 10.1111/iej.13323 [Crossref] [ Google Scholar]
- Dai X, Ma R, Jiang W, Deng Z, Chen L, Liang Y. Enterococcus faecalis-induced macrophage necroptosis promotes refractory apical periodontitis. Microbiol Spectr 2022; 10(4):e0104522. doi: 10.1128/spectrum.01045-22 [Crossref] [ Google Scholar]
- Hidalgo M, Eckhardt SG. Development of matrix metalloproteinase inhibitors in cancer therapy. J Natl Cancer Inst 2001; 93(3):178-93. doi: 10.1093/jnci/93.3.178 [Crossref] [ Google Scholar]
- Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 2002; 295(5564):2387-92. doi: 10.1126/science.1067100 [Crossref] [ Google Scholar]
- Morris GM, Lim-Wilby M. Molecular docking. Methods Mol Biol 2008; 443:365-82. doi: 10.1007/978-1-59745-177-2_19 [Crossref] [ Google Scholar]
- Bender BJ, Gahbauer S, Luttens A, Lyu J, Webb CM, Stein RM. A practical guide to large-scale docking. Nat Protoc 2021; 16(10):4799-832. doi: 10.1038/s41596-021-00597-z [Crossref] [ Google Scholar]
- Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res 2021; 49(D1):D437-51. doi: 10.1093/nar/gkaa1038 [Crossref] [ Google Scholar]
- Chen CC, Herzberg O. Inhibition of beta-lactamase by clavulanate Trapped intermediates in cryocrystallographic studies. J Mol Biol 1992; 224(4):1103-13. doi: 10.1016/0022-2836(92)90472-v [Crossref] [ Google Scholar]
- Masumi M, Noormohammadi F, Kianisaba F, Nouri F, Taheri M, Taherkhani A. Methicillin-resistant Staphylococcus aureus: docking-based virtual screening and molecular dynamics simulations to identify potential penicillin-binding protein 2A inhibitors from natural flavonoids. Int J Microbiol 2022; 2022:9130700. doi: 10.1155/2022/9130700 [Crossref] [ Google Scholar]
- Moradkhani S, Farmani A, Saidijam M, Taherkhani A. COVID-19: docking-based virtual screening and molecular dynamics study to identify potential SARS-CoV-2 spike protein inhibitors from plant-based phenolic compounds. Acta Virol 2021; 65(3):288-302. doi: 10.4149/av_2021_308 [Crossref] [ Google Scholar]
- Khoshbin E, Rahmani-Abidar SM, Moradi S, Taherkhani A, Karkehabadi H. Structure-based virtual screening of cinnamic acid analogs against RIPK3: implications for anti-inflammatory drug discovery. Avicenna J Med Biochem 2023; 11(2):129-37. doi: 10.34172/ajmb.2446 [Crossref] [ Google Scholar]
- Márton IJ, Kiss C. Protective and destructive immune reactions in apical periodontitis. Oral Microbiol Immunol 2000; 15(3):139-50. doi: 10.1034/j.1399-302x.2000.150301.x [Crossref] [ Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol 2005; 1(2):112-9. doi: 10.1038/nchembio711 [Crossref] [ Google Scholar]
- Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ 2018; 25(3):486-541. doi: 10.1038/s41418-017-0012-4 [Crossref] [ Google Scholar]
- Liu S, Joshi K, Denning MF, Zhang J. RIPK3 signaling and its role in the pathogenesis of cancers. Cell Mol Life Sci 2021; 78(23):7199-217. doi: 10.1007/s00018-021-03947-y [Crossref] [ Google Scholar]
- Orozco S, Oberst A. RIPK3 in cell death and inflammation: the good, the bad, and the ugly. Immunol Rev 2017; 277(1):102-12. doi: 10.1111/imr.12536 [Crossref] [ Google Scholar]
- Shlomovitz I, Zargrian S, Gerlic M. Mechanisms of RIPK3-induced inflammation. Immunol Cell Biol 2017; 95(2):166-72. doi: 10.1038/icb.2016.124 [Crossref] [ Google Scholar]
- Bouchouka E, Djilani A, Bekkouche A. Antibacterial and antioxidant activities of three endemic plants from Algerian Sahara. Acta Sci Pol Technol Aliment 2012; 11(1):61-5. [ Google Scholar]
- Lam KY, Ling AP, Koh RY, Wong YP, Say YH. A review on medicinal properties of orientin. Adv Pharmacol Sci 2016; 2016:4104595. doi: 10.1155/2016/4104595 [Crossref] [ Google Scholar]
- Kim SJ, Pham TH, Bak Y, Ryu HW, Oh SR, Yoon DY. Orientin inhibits invasion by suppressing MMP-9 and IL-8 expression via the PKCα/ ERK/AP-1/STAT3-mediated signaling pathways in TPA-treated MCF-7 breast cancer cells. Phytomedicine 2018; 50:35-42. doi: 10.1016/j.phymed.2018.09.172 [Crossref] [ Google Scholar]
- Tian F, Tong M, Li Z, Huang W, Jin Y, Cao Q. The effects of orientin on proliferation and apoptosis of T24 human bladder carcinoma cells occurs through the inhibition of nuclear factor-kappaB and the hedgehog signaling pathway. Med Sci Monit 2019; 25:9547-54. doi: 10.12659/msm.919203 [Crossref] [ Google Scholar]
- Xiao Q, Cui Y, Zhao Y, Liu L, Wang H, Yang L. Orientin relieves lipopolysaccharide-induced acute lung injury in mice: the involvement of its anti-inflammatory and anti-oxidant properties. Int Immunopharmacol 2021; 90:107189. doi: 10.1016/j.intimp.2020.107189 [Crossref] [ Google Scholar]
- Saha R, Patkar S, Pillai MM, Tayalia P. Bilayered skin substitute incorporating rutin nanoparticles for antioxidant, anti-inflammatory, and anti-fibrotic effect. Biomater Adv 2023; 150:213432. doi: 10.1016/j.bioadv.2023.213432 [Crossref] [ Google Scholar]
- Hamad RS. Rutin, a flavonoid compound derived from garlic, as a potential immunomodulatory and anti-inflammatory agent against murine Schistosomiasis mansoni. Nutrients 2023; 15(5):1206. doi: 10.3390/nu15051206 [Crossref] [ Google Scholar]