Avicenna J Dent Res. 14(3):130-136.
doi: 10.34172/ajdr.2022.24
Review Article
Expression of Different Tumor Biomarkers in Oral Lichen Planus: A Meta-analysis
Alberto Rodriguez-Archilla 1, *  , Benayga Herrera-Plasencia 2
, Benayga Herrera-Plasencia 2 
Author information:
1Professor of Oral Medicine, Department of Stomatology, Oral Medicine Unit, University of Granada, Granada, Spain
2Research Fellow in Oral Medicine, Department of Stomatology, Oral Medicine Unit, University of Granada, Granada, Spain
*
Corresponding author: Alberto Rodriguez-Archilla, Department of Stomatology, Oral Medicine Unit, Faculty of Dentistry, University of Granada, Granada, Spain. Tel: +34 958 244 085, +34 656 474 473, Email:
alberodr@ugr.es
Abstract
Background: Oral lichen planus (OLP) is a potentially malignant oral disorder that affects 0.5-2% of the general population with a malignant transformation rate of around 1.1%. Malignant transformation is characterized by the increased proliferation of basal layer cells under the influence of biomarkers released from the inflammatory infiltrate. This study was conducted to assess the expression of biomarkers in OLP and their possible predictive value for malignant transformation of these lesions.
Methods: A search for studies on tumor biomarkers in OLP was performed in the following databases: PubMed (MEDLINE, Cochrane Library), Web of Science, and Scopus. Data were analyzed using the statistical software RevMan 5.4 (The Cochrane Collaboration, Oxford, UK). For continuous outcomes, the estimates of effects of an intervention were expressed as mean differences (MD) using the inverse variance (IV) method, and for dichotomous outcomes, the estimates of effects of an intervention were expressed as odds ratios (OR) using Mantel-Haenszel (M-H) method, all with 95% confidence intervals.
Results: A total of 30 studies were included in this meta-analysis. OLP patients compared to controls without the disease had a significantly higher expression of mutated p53 protein (P<0.001), Ki-67 antigen (P<0.001), p16 protein (P<0.001), and cell proliferation nuclear antigen (PCNA) (P=0.04), but not blc-2 protein. In contrast, OLP patients showed 3.71 times higher probability of bcl-2 protein detection (P=0.01).
Conclusions: The expression of tumor biomarkers in OLP suggests the potentially malignant nature of some of these lesions
Keywords: Biomarkers, Tumor, Lichen planus, Oral, Mouth, Precancerous conditions
Copyright and License Information
© 2022 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Rodriguez-Archilla A, Herrera-Plasencia B. Expression of different tumor biomarkers in oral lichen planus: a meta-analysis. Avicenna J Dent Res. 2022; 14(3):130-136. doi:10.34172/ajdr.2022.24
Introduction
Oral lichen planus (OLP) is a potentially malignant oral disorder characterized by a chronic mucocutaneous inflammatory disease that affects 0.5%-2% of the general population. The age of onset is generally between 30 and 60 years, with greater involvement in the female gender (1). Clinically, OLP has multiple presentations ranging from asymptomatic white keratotic lesions to painful erosions and ulcerations with six distinctive clinical forms including white ones (reticular, papular, plaque) and red ones (erosive, atrophic, and bullous). Histopathologically, it is characterized by hydropic degeneration of basal epithelial cells and a “band” chorionic inflammatory infiltrate (2).
According to a recent study, the malignancy rate of OLP is around 1.1%, with a higher incidence in smokers, drinkers, those infected with the hepatitis C virus, and patients with erosive lesions (3). The average time of transformation of an OLP into an oral squamous cell carcinoma (OSCC) is about 5.5 years (4). OLP patients require periodic monitoring to identify early clinical and/or histopathological signs of malignant transformation to OSCC. Several biomarkers related to OLP have been studied, such as apoptosis modulators (p53 and bcl-2), cell cycle regulators (Ki-67, p16, and PCNA), tissue remodeling factors (MMPs), and inflammatory factors (TNF-α, IL-6, and COX-2). Malignant transformation is characterized by increased proliferation of basal layer cells under the influence of biomarkers released from the inflammatory infiltrate that activate different pathways and can lead to tumor development (5). This study aimed to assess the expression of tumor biomarkers in OLP and its possible predictive value for malignant transformation of these lesions.
Materials and Methods
The two authors (ARA and BHP) independently carried out all research steps (search, study selection, data extraction, and evaluation). Subsequently, they jointly agreed on the articles to include in this study.
A search for studies on tumor biomarkers and OLP up to October 2021 was performed in the following databases: PubMed (MEDLINE, Cochrane Library), Web of Science (WoS), and Scopus. Search strategies were developed for each database combining Medical Subjects Headings (MeSH) and free-text terms. The search terms were as follows: (“ki 67 antigen”[MeSH Terms] OR “tumor suppressor protein p53”[MeSH Terms] OR “proliferating cell nuclear antigen”[MeSH Terms] OR “genes, p16”[MeSH Terms] OR “bcl-2” OR “biomarkers, tumor”[MeSH Terms]) AND “lichen planus, oral”[MeSH Terms]; (“ki 67” OR “p53” OR “PCNA” OR “bcl-2”) AND “oral lichen planus”; TITLE-ABS-KEY ((“ki 67” OR “p53” OR “PCNA” OR “bcl-2”) AND “oral lichen planus”). Articles with a relevant risk of bias (score <6 points based on the Newcastle-Ottawa methodological quality assessment scale) (6), articles without full-text availability, articles without a healthy control group, studies without clinical data, and studies with non-usable data were excluded from the study.
Assessment of Methodological Quality
The methodological quality of the studies included in this manuscript was determined using the Newcastle-Ottawa methodological quality assessment scale consisting of eight items that evaluate three dimensions (selection, comparability, and exposure) (6). According to the score obtained, the studies are classified as high quality (≥7 points), moderate quality (4-6 points), and low quality (1-3 points).
Statistical Analysis
For the meta-analysis, the data were processed using RevMan 5.4 software (The Cochrane Collaboration, Oxford, UK). For continuous outcomes, the inverse of the variance (IV) for the mean difference (MD) was used, and for dichotomous outcomes, the odds ratio (OR) with the Mantel-Haenszel chi-square formula (M-H) was used, both with 95% confidence intervals (95% CI). Heterogeneity was determined according to the Higgins statistic (I2). In case of high heterogeneity (I2>50%), the random-effects model was applied. P<0.05 was considered the minimum level of significance. In addition, the MedCalc Statistical Software version 20.019 (MedCalc Software Ltd. Ostend, Belgium) was used to estimate publication bias through funnel plots and the Egger test, with a significance value of P<0.1.
Results
In the initial search, 408 articles were found (125 in PubMed, 166 in WoS, and 117 in Scopus), 122 of which were duplicates, leaving 286 articles for eligibility. There were no restrictions on language or publication date. The exclusion criteria were: (a) articles with a relevant risk of bias (<6 points) according to the Newcastle-Ottawa methodological quality assessment scale (6) (n=82), (b) articles without full-text availability (n=61), (c) articles without a healthy control group (n=46), (d) studies without clinical data (n=14), and (e) studies with non-usable data (n=53). Finally, 30 articles were included in this meta-analysis (Figure 1).
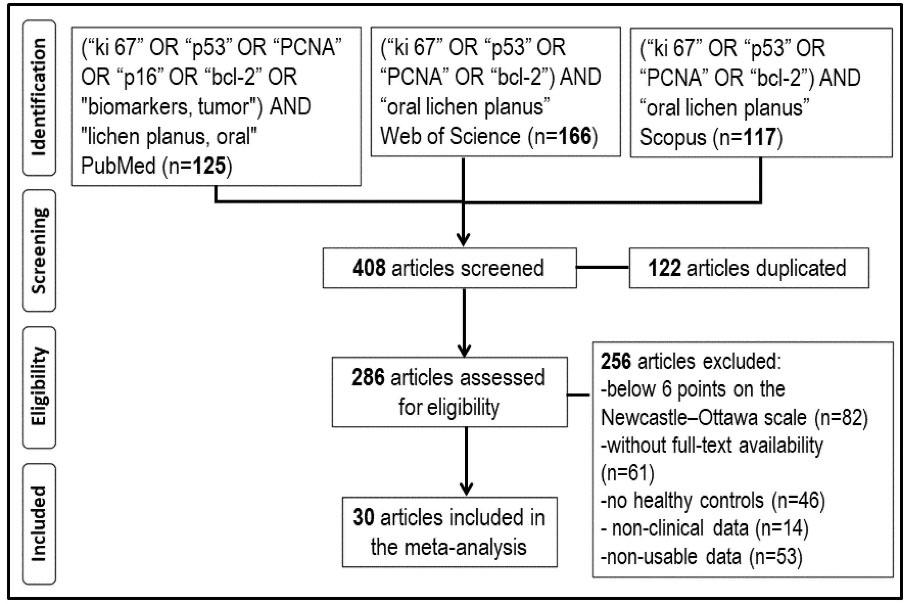
Figure 1.
Study Selection Flowchart.
.
Study Selection Flowchart.
Table 1 presents the main reporting characteristics and the methodological quality of the 30 studies included in this meta-analysis according to the NOS scale (7-36). A total of 1247 individuals, 831 patients (66.6%) with OLP, and 416 (33.4%) healthy controls were included in these articles. Among OLP patients, 62.8% were female and 37.2% were male, and among controls without the disease, 52.2% were female and 47.8% were male. Considering the Newcastle-Ottawa (NOS) quality scale (6), 20 articles (66.7%) had 6 points, 9 articles (30.0%) got 7 points, and 1 article (3.3%) reached 8 points.
Table 1.
Reporting Characteristics and Methodological Quality Evaluation of the 30 Studies Included in this Meta-analysis
|
Study
|
Year
|
Country
|
Study Population
|
Detection Method
|
Tumor Biomarker
|
NOS
|
| Schifter (7) |
1998 |
Australia |
46 OLP (12M, 34F, 56y)
18 Cont (9M, 9F, 59y) |
IMH |
p53 |
7 |
| Bloor (8) |
1999 |
UK |
36 OLP (na, na, na)
8 Cont (na, na, na) |
IMH |
Ki-67, bcl-2 |
6 |
| da Silva Fonseca (9) |
2001 |
Brazil |
20 OLP (7M, 13F, 43y)
20 Cont (7M, 13F, 43y) |
IMH |
PCNA |
6 |
| Garcia-Pola (10) |
2001 |
Spain |
10 OLP (4M, 6F, 58y)
10 Cont (5M, 5F, 56y) |
IMH |
Ki-67 |
6 |
| Valente (11) |
2001 |
Italy |
15 OLP (10M, 5F, 57y)
7 Cont (na, na, na) |
IMH |
p53 |
6 |
| Hirota (12) |
2002 |
Japan |
19 OLP (6M, 13F, 57y)
10 Cont (na, na, na) |
IMH |
Ki-67, p53, p21 |
6 |
| Ogmundsdottir (13) |
2002 |
Iceland |
48 OLP (na, na, na)
10 Cont (na, na, na) |
IMH |
p53 |
7 |
| Sklavounou-Andrikopoulou (14) |
2004 |
Greece |
26 OLP (8M, 18F, 57y)
26 Cont (11M, 15F, 47y) |
ELISA |
bcl-2 |
7 |
| Lee (15) |
2005 |
Taiwan |
56 OLP (26M, 30F, 48y)
20 Cont (na, na, na) |
IMH |
PCNA, p53 |
7 |
| González-Moles (16) |
2006 |
Spain |
51 OLP (18M, 33F, 55y)
26 Cont (13M, 13F, 55y) |
IMH |
Ki-67, p53, p21, bcl-2 |
7 |
| Montebugnoli (17) |
2006 |
Italy |
30 OLP (13M, 17F, 53y)
9 Cont (4M, 5F, 53y) |
IMH |
Ki-67, p53 |
6 |
| Abdel-Latif (18) |
2009 |
Egypt |
25 OLP (16M, 9F, 45y)
10 Cont (na, na, na) |
IMH |
bcl-2 |
6 |
| Agha-Hosseini (19) |
2009 |
Iran |
44 OLP (17M, 27F, 46y)
30 Cont (12M, 18F, 44y) |
IMH |
Ki-67, p53 |
7 |
| Freitas (20) |
2010 |
Brazil |
7 OLP (na, na, na)
7 Cont (na, na, na) |
IMH |
PCNA, p53, bcl-2 |
6 |
| Safadi (21) |
2010 |
Jordan |
18 OLP (8M, 10F, 48y)
10 Cont (5M, 5F, 47y) |
IMH |
p53, p21 |
7 |
| Poomsawat (22) |
2011 |
Thailand |
23 OLP (6M, 17F, na)
10 Cont (4M, 6F, na) |
IMH |
p16 |
6 |
| Leyva-Huerta (23) |
2012 |
Mexico |
21 OLP (5M, 16F, 56y)
4 Cont (na, na, na) |
IMH |
p53, bcl-2 |
6 |
| Dang (24) |
2013 |
China |
20 OLP (8M, 12F, 49y)
10 Cont (5M, 5F, 28y) |
PCR |
p16 |
6 |
| Zargaran (25) |
2013 |
Iran |
16 OLP (1M, 15F, 38y)
17 Cont (13M, 4F, 46y) |
IMH |
Ki-67 |
6 |
| Al-Azzawi (26) |
2014 |
Iraq |
21 OLP (na, na, na)
10 Cont (na, na, na) |
IMH |
PCNA, p53 |
6 |
| Salehinejad (27) |
2014 |
Iran |
15 OLP (na, na, na)
8 Cont (na, na, na) |
IMH |
p16 |
6 |
| Agha-Hosseini (28) |
2015 |
Iran |
34 OLP (15M, 19F, na)
41 Cont (19M, 22F, na) |
ELISA |
p53 |
7 |
| Kumar (29) |
2015 |
India |
20 OLP (na, na, na)
20 Cont (na, na, na) |
IMH |
Ki-67 |
6 |
| Pigatti (30) |
2015 |
Brazil |
14 OLP (na, na, na)
9 Cont (na, na, na) |
IMH |
Ki-67, bcl-2 |
6 |
| Basheer (31) |
2017 |
India |
10 OLP (na, na, na)
10 Cont (na, na, na) |
IMH |
p53 |
6 |
| Hadzi-Mihailovic (32) |
2017 |
Serbia |
40 OLP (12M, 28F, 58y)
13 Cont (6M, 7F, na) |
IMH |
p53 |
7 |
| Danielsson (33) |
2018 |
Sweden |
79 OLP (26M, 54F, 57y)
15 Cont (9M, 6F, 46y) |
IMH |
p16 |
8 |
| Shimada (34) |
2018 |
Japan |
20 OLP (na, na, 50y)
5 Cont (na, na, 60y) |
IMH |
Ki-67 |
6 |
| Shiva (35) |
2018 |
Iran |
32 OLP (15M, 17F, 46y)
8 Cont (na, na, na) |
IMH |
p53 |
6 |
| Beevi (36) |
2019 |
India |
15 OLP (na, na, na)
15 Cont (na, na, na) |
IMH |
Ki-67 |
6 |
OLP: oral lichen planus patients; Cont: healthy controls; M: male; F: female; y: mean age in years; na: not available; IMH: immunohistochemistry; ELISA: enzyme-linked immunosorbent assay; PCR: polymerase chain reaction; PCNA: proliferating cell nuclear antigen; NOS: Newcastle-Ottawa methodological quality scale
The analysis of various tumor biomarkers concentrations in OLP patients versus. controls without the disease is shown in Table 2.
Table 2.
Analysis of Various Tumor Biomarkers Concentrations in Oral Lichen Planus Patients vs. Controls Without the Disease
|
Tumor Biomarker
|
References
|
Value
|
MD
|
(95% CI)
|
I2(%)
|
P Value
|
| p53 |
(11,12,17,19-21,26,28,35) |
OLP |
14.17 |
(7.96 to 20.39) |
96% |
<0.001* |
| Ki-67 |
(8,10-12,17,19,25,29,36) |
OLP |
17.32 |
(11.37 to 23.27) |
95% |
<0.001* |
| PCNA |
(9,15,20,26) |
OLP |
13.98 |
(0.75 to 27.22) |
98% |
0.040* |
| bcl-2 |
(14,18,20) |
OLP |
0.02 |
(-0.04 to 0.07) |
0% |
0.560 |
PCNA: proliferating cell nuclear antigen; MD: mean difference; CI: confidence interval; I2(%): Higgins statistic for heterogeneity (percentage);
*Statistically significant.
Nine studies (11,12,17,19-21,26,28,35) analyzed the percentage of positivity for p53, indicating that OLP patients had 14.17% higher positivity, with a highly statistically significant relationship (MD=14.17; 95% CI: 7.96 to 20.39; P<0.001).
Nine studies (8,10-12,17,19,25,29,36)examined the percentage of Ki-67 antigen-positive cells. Positivity for Ki-67 increased by 17.32% in OLP patients, with highly statistically significant differences (MD=17.32; 95% CI: 11.37 to 23.27; P<0.001).
Four studies (9,15,20,26) quantified the percentage of proliferating cell nuclear antigen (PCNA) positivity, with 13.98% higher PCNA positivity in OLP patients and a statistically significant relationship (MD=13.98; 95% CI%: 0.75 to 27.22; P=0.040).
Three studies (14,18,20) determined the bcl-2 protein levels, without observing variations between the two population groups. The statistical analysis did not show significant differences (MD=0.02; 95% CI: -0.04 to 0.07; P=0.560).
Table 3 exhibits the odds ratios for several tumor biomarkers expression in subjects with and without OLP.
Table 3.
Odds Ratios and 95% Confidence Intervals for Several Tumor Biomarkers Expression in Subjects With and Without Oral Lichen Planus
|
Tumor biomarker
|
References
|
Value
|
OR
|
(95% CI)
|
I2(%)
|
P Value
|
| p53 |
(7,13,15,16,23,31,32,35) |
OLP |
5.73 |
(3.44 to 9.54) |
0% |
<0.001* |
| Ki-67 |
(15,30,34) |
OLP |
10.59 |
(1.23 to 91.33) |
70% |
0.030* |
| p16 |
(22,24,27,33) |
OLP |
18.48 |
(4.87 to 70.03) |
0% |
<0.001* |
| bcl-2 |
(16,18,30) |
OLP |
3.71 |
(1.29 to 10.65) |
0% |
0.010* |
PCNA: proliferating cell nuclear antigen; OR: odds ratio; CI: Confidence interval; I2(%): Higgins statistic for heterogeneity (percentage); *Statistically significant.
Eight studies (7,13,15,16,23,31,32,35)evaluated the p53 protein expression in 2 groups, finding that OLP patients were 5.73 times more likely to express p53 compared to controls, with a highly statistically significant association (OR=5.73; 95% CI: 3.44 to 9.54; P<0.001).
Three other studies (15,30,34)investigated the Ki-67 antigen expression, showing that OLP patients were 10.59 times more likely to express Ki-67 in their lesions. In the statistical analysis, a significant relationship was found (OR=10.59; 95% CI: 1.23 to 91.33; P=0.030).
Four studies (22,24,27,33)estimated the p16 protein expression, indicating an increase of 18.48-fold in the likelihood of p16 expression in OLP patients compared to controls. Based on statistical analysis, highly significant differences were found (OR=18.48; 95% CI: 4.87 to 70.03; P<0.001).
Three other studies (16,18,30)assessed the bcl-2 protein expression, verifying a 3.71-fold increase in the probability of blc-2 expression in OLP patients, with a highly statistically significant association (OR=3.71; 95% CI: 1.29 to 10.65; P=0.010).
The evaluation of publication bias for p53 protein, Ki-67 antigen, bcl-2 protein, and PCNA is shown in Figure 2. The visual inspection of funnel plots displays some asymmetry for p53 and PCNA with probable publication bias, but not for Ki-67 and bcl-2. At the same time, based on the results of Egger’s test, there was some publication bias for p53 (t=7.80, P=0.04) and PCNA (t=-8.24, P=0.02); however, no publication bias was detected for Ki-67 (t=5.96, P=0.13) and bcl-2 (t=-1.59, P=0.34).
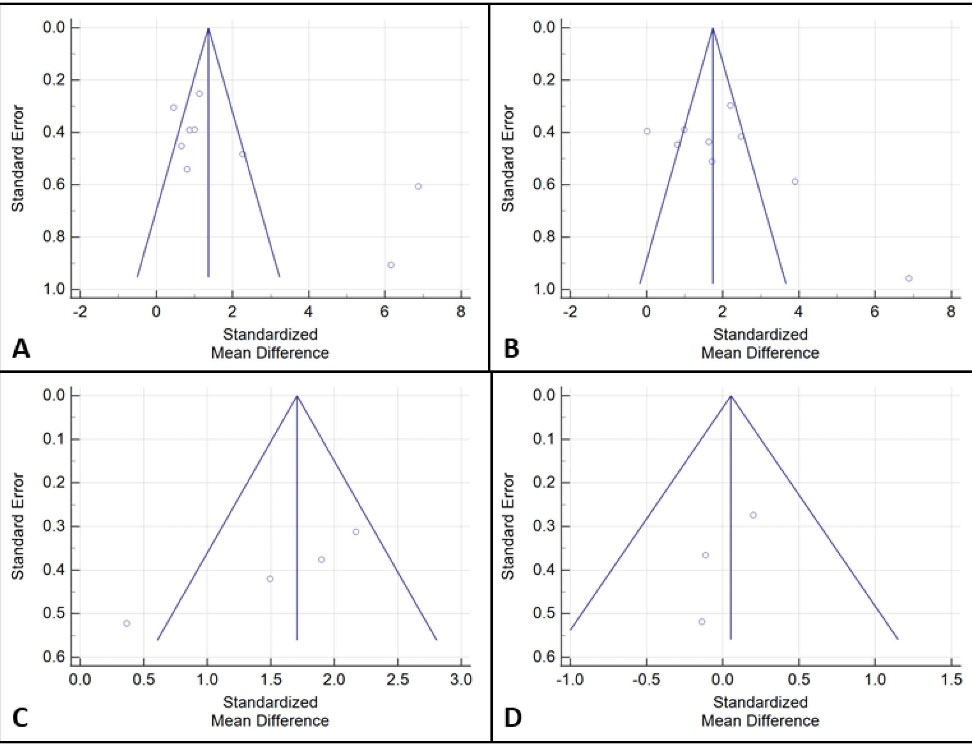
Figure 2.
Funnel Plots for Publication Bias Assessment of the Levels of p53 Protein (A), Ki-67 Antigen (B), bcl-2 Protein (C), and PCNA (D) in OLP Patients
.
Funnel Plots for Publication Bias Assessment of the Levels of p53 Protein (A), Ki-67 Antigen (B), bcl-2 Protein (C), and PCNA (D) in OLP Patients
Discussion
In the present meta-analysis on the expression of tumor biomarkers in OLP, data from 30 studies were included.
The TP53 gene, called the “genome guardian”, encodes the tumor suppressor protein p53. This protein performs important functions such as the arrest of cell division (senescence), the induction of programmed cell death (apoptosis), or the DNA repair in mutated cells. Protein p53 activation after DNA damage or oncogenic signaling is an important protective mechanism, facilitating DNA repair and stimulating the apoptosis of damaged cells. Functional loss and altered expression of the p53 protein are the most common genetic changes (mutations) found in human cancers (28).
In this study, OLP patients showed 14.17% higher positivity for p53 than controls, with a highly statistically significant relationship (P<0.001). Nine studies (11,12,17,19-21,26,28,35) that evaluated the percentage of p53 positivity indicated the highest percentage of expression of this protein in OLP patients. Likewise, these patients were 5.73 times more likely to express p53 compared to controls, with a highly statistically significant association (P<0.001). All the studies (7,13,15,16,23,31,32,35) that analyzed this protein confirmed this higher detection of p53 in cases of OLP. This increased p53 expression may be due to the cellular immune response that develops in OLP, inducing epithelial dysplastic changes (28). González-Moles et al (16) suggested that, in OLP, p53 overexpression would act mainly by arresting the cell cycle to induce DNA repair in cells mutated in these lesions.
The Ki-67 antigen is an immunohistochemical marker of cell proliferation expressed in the G2 and M phases of cell division, being a biological marker of mitotic activity. The positivity for the Ki-67 antigen is correlated with the clinical course of the disease, providing relevant information on the worst biological behavior and the higher probability of malignancy for the lesions (36). In the present study, OLP patients presented 17.32% higher positivity for the Ki-67 antigen compared to controls. Based on statistical analysis, highly significant differences were found (P<0.001). Nine studies (8,10-12,17,19,25,29,36)that assessed positivity for Ki-67 are in agreement with this finding. It was also observed that OLP patients were 10.59 times more likely to express Ki-67 in their lesions, with a statistically significant association (P=0.030). All the studies (15,30,34) that examined the Ki-67 antigen expression confirmed this increased risk of Ki-67 detection in OLP lesions. The increase in the expression of Ki-67 antigen in OLP patients would be correlated with the proliferative activity and the degree of dysplasia of the epithelial cells, suggesting a more active biological behavior of these lesions (29).
PCNA is a cofactor of DNA polymerase-delta that participates in DNA synthesis during the S phase of the cell cycle and its detection is used to evaluate the degree of cell proliferation (26). In this study, OLP patients showed 13.98% more positivity for PCNA, indicating statistically significant differences (P=0.040). Four studies (9,15,20,26) that investigated this antigen confirmed this higher expression in OLP lesions. The increase in PCNA positivity is suggestive of an alteration in the cell differentiation mechanism and may also be related to the presence of growth factors in OLP induced by the chronic inflammation seen in this disease (26).
The p16 protein is the product of the CDKN2 gene located on chromosome 9p21 and plays a crucial role in cell cycle regulation. This protein prevents the association of CDK4/CDK6 with cyclin D which, in turn, prevents the phosphorylation of important substrates in the G1 phase of the cell cycle, resulting in the inhibition of cell proliferation. Overexpression of p16 is a common occurrence in potentially malignant and malignant oral lesions (33). In the present meta-analysis, OLP patients had 18.48 times higher risk of p16 protein detection, with a highly statistically significant relationship (P<0.001). All studies (22,24,27,33) that analyzed this protein indicated a higher detection rate of p16 in OLP lesions. The increase in the p16 protein expression could be explained by the increase in the release of pro-inflammatory cytokines such as IFN-γ or TNF-α in OLP patients since these mediators are related to increased p16 expression (33).
Several proteins of the Bcl gene family are involved in the regulation of programmed cell death either by preventing apoptosis (Bcl-2, Bcl-xL, Bcl-w, Mcl-I) or promoting apoptosis (Bax, Bik, Bak, Bad, Bcl-xs). Because many of these proteins are co-expressed in the same cells, the ratio of anti-apoptotic to pro-apoptotic proteins determines the inherent susceptibility of a given cell to respond to apoptotic signals. The absence or low rate of apoptosis observed in OLP could be the consequence of antiapoptotic actions exerted by the bcl-2 protein on basal cells (18).
OLP patients had slightly higher levels of bcl-2 protein expression than controls, although statistical significance was not reached (P=0.560). Three studies (14,18,20) that focused on bcl-2 found no conclusive results. In contrast, OLP patients had 3.71 times higher probability of expressing this protein compared to controls, with a statistically significant association (P=0.010). Three studies (16,18,30) that investigated the bcl-2 expression in OLP patients showed a higher probability of detection in these patients. The bcl-2 protein seems not to be directly involved in the epithelial changes that occur in OLP, since keratinocytes do not show immunoreactivity for this protein (14). On other occasions, bcl-2 could be conjugated with other proteins such as bax, bad, or bcl-xL, making its detection difficult (20). On the contrary, overexpression of p16 protein has been observed in the inflammatory infiltrates, inhibiting the apoptosis of the lymphocytes and promoting the band-like inflammatory cells infiltrate, distinctive of OLP (22).
This study presents some limitations. First, the results of this meta-analysis should be interpreted with caution due to the high heterogeneity observed in some comparisons. Second, some studies did not allow an adequate assessment of the clinical type of OLP or its severity. In others, there was a lack of data on the population characteristics (age and gender) or the time of disease evolution. Third, the use of different detection methods for biomarkers could have influenced the results.
New studies are required to delve into the degree of implication of these tumor biomarkers in the biological behavior and prognosis of OLP, a common potentially malignant oral disorder.
Conclusion
In this meta-analysis, OLP patients compared to controls without the disease had a significantly higher expression of the mutated p53 protein (P<0.001), Ki-67 antigen (P<0.001), p16 protein (P<0.001), and cell proliferation nuclear antigen (PCNA) (P=0.040), but not blc-2 protein. In contrast, OLP patients had 3.71 times higher probability of bcl-2 protein detection (P=0.010).
Authors’ Contribution
ARA and BHP contributed equally to the study design, data collection, data analysis, and manuscript preparation. Both approved the final version.
Conflict of Interest Disclosures
The authors declare that they have no conflict of interests.
Ethical Statement
Not applicable.
Funding
None.
References
- Eisen D, Carrozzo M, Bagan Sebastian JV, Thongprasom K. Number V Oral lichen planus: clinical features and management. Oral Dis 2005; 11(6):338-49. doi: 10.1111/j.1601-0825.2005.01142.x [Crossref] [ Google Scholar]
- Shen ZY, Liu W, Zhu LK, Feng JQ, Tang GY, Zhou ZT. A retrospective clinicopathological study on oral lichen planus and malignant transformation: analysis of 518 cases. Med Oral Patol Oral Cir Bucal 2012; 17(6):e943-7. doi: 10.4317/medoral.17778 [Crossref] [ Google Scholar]
- González-Moles M, Ruiz-Ávila I, González-Ruiz L, Ayén Á, Gil-Montoya JA, Ramos-García P. Malignant transformation risk of oral lichen planus: a systematic review and comprehensive meta-analysis. Oral Oncol 2019; 96:121-30. doi: 10.1016/j.oraloncology.2019.07.012 [Crossref] [ Google Scholar]
- Muñoz AA, Haddad RI, Woo SB, Bhattacharyya N. Behavior of oral squamous cell carcinoma in subjects with prior lichen planus. Otolaryngol Head Neck Surg 2007; 136(3):401-4. doi: 10.1016/j.otohns.2006.09.023 [Crossref] [ Google Scholar]
- Tampa M, Caruntu C, Mitran M, Mitran C, Sarbu I, Rusu LC. Markers of oral lichen planus malignant transformation. Dis Markers 2018; 2018:1959506. doi: 10.1155/2018/1959506 [Crossref] [ Google Scholar]
- Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, Tugwell P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Ottawa, Canada: The Ottawa Hospital. Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed November 4, 2021.
- Schifter M, Jones AM, Walker DM. Epithelial p53 gene expression and mutational analysis, combined with growth fraction assessment, in oral lichen planus. J Oral Pathol Med 1998; 27(7):318-24. doi: 10.1111/j.1600-0714.1998.tb01963.x [Crossref] [ Google Scholar]
- Bloor BK, Malik FK, Odell EW, Morgan PR. Quantitative assessment of apoptosis in oral lichen planus. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1999; 88(2):187-95. doi: 10.1016/s1079-2104(99)70116-2 [Crossref] [ Google Scholar]
- da Silva Fonseca LM, do Carmo MA. Identification of the AgNORs, PCNA and ck16 proteins in oral lichen planus lesions. Oral Dis 2001; 7(6):344-8. doi: 10.1034/j.1601-0825.2001.00747.x [Crossref] [ Google Scholar]
- García-Pola Vallejo MJ, Anitua Roldán MJ, Fernández Alvarez BE, Garcia Martín JM, López-Muñiz A. Study comparative of Ki-67 expression in oral lichen planus and oral leukoplakia Quantitative analysis. Med Oral 2001; 6(5):364-70. [ Google Scholar]
- Valente G, Pagano M, Carrozzo M, Carbone M, Bobba V, Palestro G. Sequential immunohistochemical p53 expression in biopsies of oral lichen planus undergoing malignant evolution. J Oral Pathol Med 2001; 30(3):135-40. doi: 10.1034/j.1600-0714.2001.300302.x [Crossref] [ Google Scholar]
- Hirota M, Ito T, Okudela K, Kawabe R, Yazawa T, Hayashi H. Cell proliferation activity and the expression of cell cycle regulatory proteins in oral lichen planus. J Oral Pathol Med 2002; 31(4):204-12. doi: 10.1034/j.1600-0714.2002.310403.x [Crossref] [ Google Scholar]
- Ogmundsdóttir HM, Hilmarsdóttir H, Astvaldsdóttir A, Jóhannsson JH, Holbrook WP. Oral lichen planus has a high rate of TP53 mutations A study of oral mucosa in icelanD. Eur J Oral Sci 2002; 110(3):192-8. doi: 10.1034/j.1600-0447.2002.21235.x [Crossref] [ Google Scholar]
- Sklavounou-Andrikopoulou A, Chrysomali E, Iakovou M, Garinis GA, Karameris A. Elevated serum levels of the apoptosis related molecules TNF-alpha, Fas/Apo-1 and Bcl-2 in oral lichen planus. J Oral Pathol Med 2004; 33(7):386-90. doi: 10.1111/j.1600-0714.2004.00221.x [Crossref] [ Google Scholar]
- Lee JJ, Kuo MY, Cheng SJ, Chiang CP, Jeng JH, Chang HH. Higher expressions of p53 and proliferating cell nuclear antigen (PCNA) in atrophic oral lichen planus and patients with areca quid chewing. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2005; 99(4):471-8. doi: 10.1016/j.tripleo.2004.10.018 [Crossref] [ Google Scholar]
- González-Moles MA, Bascones-Ilundain C, Gil Montoya JA, Ruiz-Avila I, Delgado-Rodríguez M, Bascones-Martínez A. Cell cycle regulating mechanisms in oral lichen planus: molecular bases in epithelium predisposed to malignant transformation. Arch Oral Biol 2006; 51(12):1093-103. doi: 10.1016/j.archoralbio.2006.06.007 [Crossref] [ Google Scholar]
- Montebugnoli L, Farnedi A, Marchetti C, Magrini E, Pession A, Foschini MP. High proliferative activity and chromosomal instability in oral lichen planus. Int J Oral Maxillofac Surg 2006; 35(12):1140-4. doi: 10.1016/j.ijom.2006.07.018 [Crossref] [ Google Scholar]
- Abdel-Latif AM, Abuel-Ela HA, El-Shourbagy SH. Increased caspase-3 and altered expression of apoptosis-associated proteins, Bcl-2 and Bax in lichen planus. Clin Exp Dermatol 2009; 34(3):390-5. doi: 10.1111/j.1365-2230.2008.03029.x [Crossref] [ Google Scholar]
- Agha-Hosseini F, Khalili M, Rohani B. Immunohistochemistry analysis of p53 and Ki-67 proteins in oral lichen planus and normal oral mucosa. Iran J Public Health 1970; 38(2):37-43. [ Google Scholar]
- de Almeida Freitas R, de Amorim RF, de Queiroz SB, da Costa Miguel MC, Silveira ÉJ, GODOY GP. Expression of PCNA, p53 and Bcl-2 in oral lichen planus: an immunohistochemical study regard-ing the potential of malignant transformation. Oral Sci 2010; 2:9-16. [ Google Scholar]
- Safadi RA, Al Jaber SZ, Hammad HM, Hamasha AA. Oral lichen planus shows higher expressions of tumor suppressor gene products of p53 and p21 compared to oral mucositis An immunohistochemical study. Arch Oral Biol 2010; 55(6):454-61. doi: 10.1016/j.archoralbio.2010.03.019 [Crossref] [ Google Scholar]
- Poomsawat S, Buajeeb W, Khovidhunkit SO, Punyasingh J. Overexpression of CDK4 and p16 in oral lichen planus supports the concept of premalignancy. J Oral Pathol Med 2011; 40(4):294-9. doi: 10.1111/j.1600-0714.2010.01001.x [Crossref] [ Google Scholar]
- Leyva-Huerta ER, Ledesma-Montes C, Rojo-Botello RE, Vega-Memije E. P53 and Bcl-2 immunoexpression in patients with oral lichen planus and oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal 2012; 17(5):e745-50. doi: 10.4317/medoral.18013 [Crossref] [ Google Scholar]
- Dang J, Bian YQ, Sun JY, Chen F, Dong GY, Liu Q. MicroRNA-137 promoter methylation in oral lichen planus and oral squamous cell carcinoma. J Oral Pathol Med 2013; 42(4):315-21. doi: 10.1111/jop.12012 [Crossref] [ Google Scholar]
- Zargaran M, Jamshidi S, Eshghyar N, Moghimbeigi A. Suitability/unsuitability of cell proliferation as an indicator of malignant potential in oral lichen planus: an immunohistochemical study. Asian Pac J Cancer Prev 2013; 14(11):6979-83. doi: 10.7314/apjcp.2013.14.11.6979 [Crossref] [ Google Scholar]
- Al-Azzawi LM. Immunohistochemical analysis of PCNA and p53 proteins in oral lichen planus, oral dysplasia and normal oral mucosa. Diyala J Med 2014; 6(1):41-7. [ Google Scholar]
- Salehinejad J, Sharifi N, Amirchaghmaghi M, Ghazi N, Shakeri MT, Ghazi A. Immunohistochemical expression of p16 protein in oral squamous cell carcinoma and lichen planus. Ann Diagn Pathol 2014; 18(4):210-3. doi: 10.1016/j.anndiagpath.2014.03.009 [Crossref] [ Google Scholar]
- Agha-Hosseini F, Mirzaii-Dizgah I, Miri-Zarandi N. Unstimulated salivary p53 in patients with oral lichen planus and squamous cell carcinoma. Acta Med Iran 2015; 53(7):439-43. [ Google Scholar]
- Kumar KV, Chaithanya KH, Punde P, Thorat A, Jangam AG, Deepthi S. Comparative evaluation of imunohistochemical expression of Ki-67 in oral lichen planus, oral leukoplakia and normal mucosa cases. J Int Oral Health 2015; 7(10):82-7. [ Google Scholar]
- Pigatti FM, Taveira LA, Soares CT. Immunohistochemical expression of Bcl-2 and Ki-67 in oral lichen planus and leukoplakia with different degrees of dysplasia. Int J Dermatol 2015; 54(2):150-5. doi: 10.1111/ijd.12279 [Crossref] [ Google Scholar]
- Basheer S, Shameena PM, Sudha S, Varma S, Vidyanath S, Varekar A. Expression of survivin and p53 in oral lichen planus, lichenoid reaction and lichenoid dysplasia: an immunohistochemical study. J Oral Maxillofac Pathol 2017; 21(3):456-7. doi: 10.4103/jomfp.JOMFP_39_15 [Crossref] [ Google Scholar]
- Hadzi-Mihailovic M, Petrovic R, Raybaud H, Stanimirovic D, Ozar Koray M. Expression and role of p53 in oral lichen planus patients. J BUON 2017; 22(5):1278-86. [ Google Scholar]
- Danielsson K, Olah J, Zohori-Zangeneh R, Nylander E, Ebrahimi M. Increased expression of p16 in both oral and genital lichen planus. Med Oral Patol Oral Cir Bucal 2018; 23(4):e449-e53. doi: 10.4317/medoral.22432 [Crossref] [ Google Scholar]
- Shimada K, Ochiai T, Shen FC, Hasegawa H. Phenotypic alteration of basal cells in oral lichen planus; switching keratin 19 and desmoglein 1 expression. J Oral Sci 2018; 60(4):507-13. doi: 10.2334/josnusd.17-0396 [Crossref] [ Google Scholar]
- Shiva A, Zamanian A, Arab S, Boloki M. Immunohistochemical study of p53 expression in patients with erosive and non-erosive oral lichen planus. J Dent (Shiraz) 2018; 19(2):118-23. [ Google Scholar]
- Beevi BH, Nayak SR, Peter CD, Haridas AK, Jacob L, Aboobakker A. Analysis of Ki-67 expression in oral premalignant lesions and normal oral mucosa: an immunohistochemical study. J Pharm Bioallied Sci 2019; 11(Suppl 2):S232-S5. doi: 10.4103/jpbs.jpbs_305_18 [Crossref] [ Google Scholar]